Chapter Summary, Questions Answers - Protein Synthesis
| Home | | Biochemistry |Chapter: Biochemistry : Protein Synthesis
Codons are composed of three nucleotide bases presented in the messenger (mRNA) language of adenine (A), guanine (G), cytosine (C), and uracil (U).
CHAPTER SUMMARY
Codons are composed of
three nucleotide bases presented in the messenger (mRNA) language of adenine
(A), guanine (G), cytosine (C), and uracil (U). They are always written 5I →3I
. Of the 64 possible three-base combinations, 61 code for the 20 common amino
acids and 3 signal termination of protein synthesis (translation). Altering the
nucleotide sequence in a codon can cause silent mutations (the altered codon
codes for the original amino acid), missense mutations (the altered codon codes
for a different amino acid), or nonsense mutations (the altered codon is a
termination codon). Characteristics of the genetic code include specificity,
universality, and degeneracy, and it is nonoverlapping and commaless (Figure
31.17). Requirements for protein synthesis include all the amino acids that
eventually appear in the finished protein, at least one specific type of transfer
RNA (tRNA) for each amino acid, one aminoacyl-tRNA synthetase for each amino
acid, the mRNA coding for the protein to be synthesized, fully competent
ribosomes, protein factors needed for initiation, elongation, and termination
of protein synthesis, and ATP and GTP as energy sources. tRNA has an attachment
site for a specific amino acid at its 3I -end, and an anticodon region that can
recognize the codon specifying the amino acid the tRNA is carrying. Ribosomes
are large complexes of protein and ribosomal (rRNA). They consist of two
subunits. Each ribosome has three binding sites for tRNA molecules: the A, P,
and E sites that cover three neighboring codons. The A-site codon binds an
incoming aminoacyl-tRNA, the P-site codon is occupied by peptidyl-tRNA, and the
E site is occupied by the empty tRNA as it is about to exit the ribosome.
Recognition of an mRNA codon is accomplished by the tRNA anticodon. The
anticodon binds to the codon following the rules of complementarity and
antiparallel binding. (Nucleotide sequences are always assumed to be written in
the 5I to 3I direction unless otherwise noted.) The “wobble” hypothesis states
that the first (5I ) base of the anticodon is not as spatially defined as the
other two bases. Movement of that first base allows nontraditional base-pairing
with the last (3I ) base of the codon, thus allowing a single tRNA to recognize
more than one codon for a specific amino acid. For initiation of protein
synthesis, the components of the translation system are assembled, and mRNA
associates with the small ribosomal subunit. The process requires initiation
factors. In prokaryotes, a purine-rich region of the mRNA (the Shine-Dalgarno
sequence) base-pairs with a complementary sequence on 16S rRNA, resulting in
the positioning of the small subunit on the mRNA so that translation can begin.
The 5I -cap (bound by proteins of the eIF-4 family) on eukaryotic mRNA is used
to position the small subunit on the mRNA. The initiation codon is AUG, and
N-formylmethionine is the initiating amino acid in prokaryotes, whereas
methionine is used in eukaryotes. The polypeptide chain is elongated by the
addition of amino acids to the carboxyl end of its growing chain. The process
requires elongation factors that facilitate the binding of the aminoacyl-tRNA
to the A site as well as the movement of the ribosome along the mRNA. The
formation of the peptide bond is catalyzed by peptidyltransferase, which is an
activity intrinsic to the rRNA of the large subunit and, therefore, is a
ribozyme. Following peptide bond formation, the ribosome advances along the
mRNA in the 5I →3I direction to the next codon (translocation). Because of the
length of most mRNAs, more than one ribosome at a time can translate a message,
forming a polysome. Termination begins when one of the three termination codons
moves into the A site. These codons are recognized by release factors. The
newly synthesized protein is released from the ribosomal complex, and the
ribosome is dissociated from the mRNA. Initiation, elongation, and termination
are driven by the hydrolysis of GTP. Initiation in eukaryotes also requires ATP
for scanning. Numerous antibiotics interfere with the process of protein
synthesis. Many polypeptide chains are covalently modified during or after
translation. Such modifications include removal of amino acids;
phosphorylation, which may activate or inactivate the protein; glycosylation,
which plays a role in protein targeting; and hydroxylation such as that seen in
collagen. Proteins must fold to achieve their functional form. Folding can be
spontaneous or facilitated by chaperones. Proteins that are defective (for
example misfolded) or destined for rapid turnover are marked for destruction by
the attachment of chains of a small, highly conserved protein called ubiquitin.
Ubiquitinated proteins are rapidly degraded by a cytosolic complex known as the
proteasome.
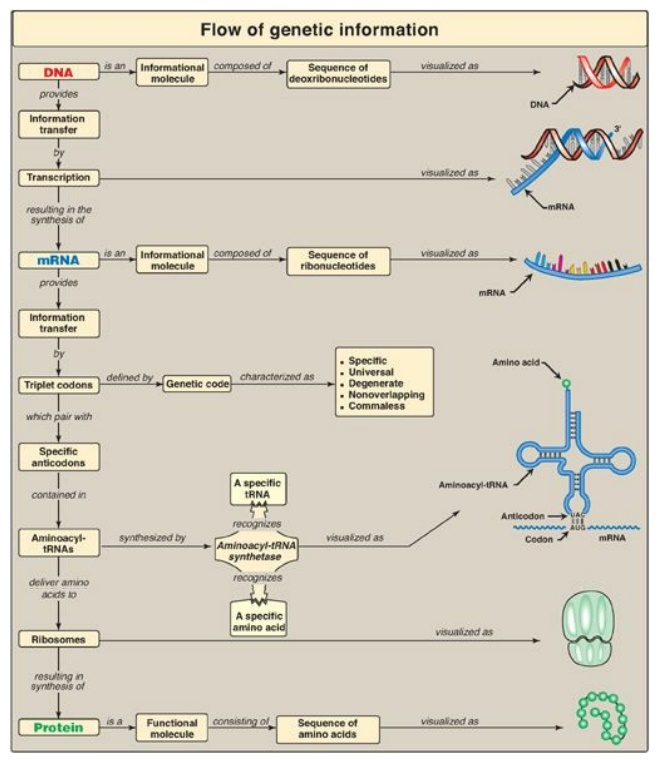
Figure 31.17 Key concept map for protein synthesis. mRNA = messenger RNA; tRNA = transfer RNA; A = adenine; G = guanine; C = cytosine; U = uracil.
Study Questions
Choose the ONE best answer.
31.1 A 20-year-old man with a microcytic anemia is
found to have an abnormal form of β-globin (Hemoglobin Constant Spring) that is
172 amino acids long, rather than the 141 found in the normal protein. Which of
the following point mutations is consistent with this abnormality?
A. CGA → UGA
B. GAU → GAC
C. GCA → GAA
D. UAA → CAA
E. UAA → UAG
Correct answer = D. Mutating the normal termination
(stop) codon for β-globin from UAA to CAA causes the ribosome to insert a
glutamine at that point. It will continue extending the protein chain until it
comes upon the next stop codon further down the message, resulting in an
abnormally long protein. The replacement of CGA (arginine) with UGA (stop)
would cause the protein to be too short. GAU and GAC both encode aspartate and
would cause no change in the protein. Changing GCA (alanine) to GAA (glutamate)
would not change the size of the protein product. A change from UAA to UAG
would simply change one termination codon for another, and would have no effect
on the protein
31.2 A pharmaceutical company is studying a new
antibiotic that inhibits bacterial protein synthesis. When this antibiotic is
added to an in vitro protein synthesis system that is translating the messenger
RNA sequence AUGUUUUUUUAG, the only product formed is the dipeptide fMet-Phe.
What step in protein synthesis is most likely inhibited by the antibiotic?
A. Initiation
B. Binding of charged
transfer RNA to the ribosomal A site
C. Peptidyltransferase
activity
D. Ribosomal translocation
E. Termination
Correct answer = D. Because fMet-Phe is made, the
ribosomes must be able to complete initiation, bind Phe-tRNA to the A site, and
use peptidyltransferase activity to form the first peptide bond. Because the ribosome
is not able to proceed any further, ribosomal movement (translocation) is most
likely the inhibited step. The ribosome is, therefore, frozen before it reaches
the termination codon of this message.
31.3 A transfer RNA (tRNA) molecule that is
supposed to carry cysteine (tRNAcys) is mischarged, so that it actually carries
alanine (ala-tRNAcys). Assuming no correction occurs, what will be the fate of
this alanine residue during protein synthesis?
A. It will be
incorporated into a protein in response to a codon for alanine.
B. It will be incorporated into a protein in
response to a codon for cysteine.
C. It will be
incorporated randomly at any codon.
D. It will remain
attached to the tRNA because it cannot be used for protein synthesis.
E. It will be
chemically converted to cysteine by cellular enzymes.
Correct answer = B. Once an amino acid is attached to
a transfer (tRNA) molecule, only the anticodon of that tRNA determines the
specificity of incorporation. The mischarged alanine will, therefore, be
incorporated into the protein at a position determined by a cysteine codon.
31.4 In a patient with cystic fibrosis caused by
the ∆F508 mutation, the mutant cystic fibrosis transmembrane conductance
regulator (CFTR) protein folds incorrectly. The patient s cells modify this
abnormal protein by attaching ubiquitin molecules to it. What is the fate of
this modified CFTR protein?
A. It performs its
normal function because the ubiquitin largely corrects for the effect of the
mutation.
B. It is secreted from
the cell.
C. It is placed into
storage vesicles.
D. It is degraded by the proteasome.
E. It is repaired by
cellular enzymes.
Correct answer = D. Ubiquitination usually marks old,
damaged, or misfolded proteins for destruction by the cytosolic proteasome.
There is no known cellular mechanism for repair of damaged proteins.
31.5 Many antimicrobials inhibit protein
translation. Which of the following antimicrobials is correctly paired with its
mechanism of action?
A. Erythromycin binds
to the 60S ribosomal subunit.
B. Puromycin
inactivates EF-2.
C. Streptomycin binds to the 30S ribosomal subunit.
D. Tetracyclines
inhibit peptidyltransferase.
Correct answer = C. Streptomycin binds the 30S subunit
and inhibits translation initiation. Erythromycin binds the 50S ribosomal
subunit (60S denotes a eukaryote) and blocks the tunnel through which the
peptide leaves the ribosome. Puromycin has structural similarity to
aminoacyl-tRNA. It is incorporated into the growing chain, inhibits elongation,
and results in premature termination in both prokaryotes and eukaryotes.
Tetracyclines bind the 30S ribosomal subunit and block access to the A site,
inhibiting elongation.
31.6 Translation of a synthetic polyribonucleotide
containing the repeating sequence CAA in a cell-free protein-synthesizing
system produces three homopolypeptides: polyglutamine, polyasparagine, and
polythreonine. If the codons for glutamine and asparagine are CAA and AAC,
respectively, which of the following triplets is the codon for threonine?
A. AAC
B. ACA
C. CAA
D. CAC
E. CCA
Correct answer = B. The synthetic polynucleotide
sequence of CAACAACAACAA.. could be read by the in vitro protein synthesizing
system starting at the first C, the first A, or the second A. In the first
case, the first triplet codon would be CAA, which codes glutamine; in the
second case, the first triplet codon would be AAC, which codes for asparagine;
in the last case, the first triplet codon would be ACA, which codes for
threonine.
31.7 Which of the following is required for both
prokaryotic and eukaryotic protein synthesis?
A. Binding of the small
ribosomal subunit to the Shine-Dalgarno sequence
B. fMet-tRNA
C. Movement of the
messenger RNA out of the nucleus and into the cytoplasm
D. Recognition of the 5
-cap by initiation factors.
E. Translocation of the peptidyl-tRNA from the A
site to the P site
Correct answer = E. In both prokaryotes and
eukaryotes, continued translation (elongation) requires movement of the
peptidyl-tRNA from the A to the P site to allow the next aminoacyl-tRNA to
enter the A site. Only prokaryotes have a Shine-Dalgarno sequence and use fMet,
and only eukaryotes have a nucleus and co- and posttranscriptionally process
their mRNA.
31. 8 α1-Antitrypsin (AAT) deficiency can result in
emphysema, a lung pathology, because the action of elastase, a serine protease,
is unopposed. Deficiency of AAT in the lungs is the consequence of impaired
secretion from the liver, the site of its synthesis. Proteins such as AAT that
are destined to be secreted are best characterized by which of the following
statements?
A. Their synthesis is
initiated on the smooth endoplasmic reticulum.
B. They contain a
mannose 6-phosphate targeting signal.
C. They always contain
methionine as the N-terminal amino acid.
D. They are produced from translation products that
have an N-terminal hydrophobic signal sequence.
E. They contain no
sugars with O-glycosidic linkages because their synthesis does not involve the
Golgi apparatus.
Correct answer = D. Synthesis of secreted proteins is
begun on free (cytosolic) ribosomes. As the N-terminal signal sequence of the
peptide emerges from the ribosome, it is bound by the signal recognition
particle, taken to the rough endoplasmic reticulum (RER), threaded into the
lumen, and removed as translation continues. The proteins move through the RER
and the Golgi, and undergo processing such as N-glycosylation (RER) and
O-glycosylation (Golgi). In the Golgi, they are packaged in secretory vesicles
and released from the cell. The smooth endoplasmic reticulum is associated with
synthesis of lipids, not proteins, and has no ribosomes attached.
Phosphorylation at carbon 6 of terminal mannose residues in glycoproteins
targets these proteins (acid hydrolases) to lysosomes. The N-terminal
methionine is removed from most proteins during processing.
31.9 Why is the genetic code described both as
degenerate and unambiguous?
A given amino acid can
be coded for by more than one codon (degenerate code), but a given codon codes
for just one particular amino acid (unambiguous code).
Related Topics
