Chapter Summary, Questions Answers - RNA Structure, Synthesis, and Processing
| Home | | Biochemistry |Chapter: Biochemistry : RNA Structure, Synthesis, and Processing
There are three major types of RNA that participate in the process of protein synthesis: ribosomal RNA (rRNA), transfer RNA (tRNA), and messenger RNA (mRNA).
CHAPTER SUMMARY
There are three major types of RNA that participate in the process of protein synthesis: ribosomal RNA (rRNA), transfer RNA (tRNA), and messenger RNA (mRNA) (Figure 30.20). They are unbranched polymers of nucleotides but differ from DNA by containing ribose instead of deoxyribose and uracil instead of thymine. rRNA is a component of the ribosomes. tRNA serves as an “adaptor” molecule that carries a specific amino acid to the site of protein synthesis. mRNA carries genetic information from DNA for use in protein synthesis. The process of RNA synthesis is called transcription, and the substrates are ribonucleoside triphosphates. The enzyme that synthesizes RNA is RNA polymerase (RNA pol). In prokaryotic cells, the core enzyme has five subunits (2α, 1β, 1β , and 1Ω) and possesses 5I →3I polymerase activity that elongates the growing RNA strand. This enzyme requires an additional subunit, sigma (s) factor, that recognizes the nucleotide sequence (promoter region) at the beginning of a length of DNA that is to be transcribed. This region contains consensus sequences that are highly conserved and include the TATA (Pribnow) box and the –35 sequence. Another protein—rho (r) factor—is required for termination of transcription of some genes. There are three distinct classes of RNA pol in the nucleus of eukaryotic cells. RNA pol I synthesizes the precursor of rRNAs in the nucleolus. In the nucleoplasm, RNA pol II synthesizes the precursors for mRNA and some noncoding RNAs, and RNA pol III produces the precursors of tRNA. In both prokaryotes and eukaryotes, RNA pol does not require a primer and has no 3I →5I exonuclease (proofreading) activity. Core promoters for genes transcribed by RNA pol II contain cis-acting consensus sequences, such as the TATA-like Hogness box, that serve as binding sites for trans-acting general transcription factors. Upstream of these are proximal regulatory elements, such as the CAAT and GC boxes, and distal regulatory elements such as enhancers. Transcriptional activators (specific transcription factors) bind these elements and regulate the frequency of transcription initiation, the response to signals such as hormones, and which genes are expressed at a given point in time. Eukaryotic transcription requires that the chromatin be accessible. A primary transcript is a linear copy of a transcription unit, the segment of DNA between specific initiation and termination sequences. The primary transcripts of both prokaryotic and eukaryotic tRNA and rRNA are posttranscriptionally modified by cleavage of the original transcripts by ribonucleases. rRNAs of both prokaryotic and eukaryotic cells are synthesized from long precursor molecules called pre-rRNA. These precursors are cleaved and trimmed by ribonucleases, producing the three largest rRNA, and bases and sugars are modified. Eukaryotic 5S rRNA is synthesized by RNA pol III and is modified separately. Prokaryotic mRNA is generally identical to its primary transcript, whereas eukaryotic mRNA is extensively modified co- and posttranscriptionally. For example, a 7-methylguanosine cap is attached to the 5I -terminal end of the mRNA through a 5I →5I linkage. A long poly-A tail, not transcribed from the DNA, is attached to the 3I -end of most mRNAs. Most eukaryotic mRNAs also contain intervening sequences (introns) that must be removed to make the mRNA functional. Their removal, as well as the joining of expressed sequences (exons), requires a spliceosome composed of small, nuclear ribonucleoprotein particles (snurps) that mediate the process of splicing. Eukaryotic mRNA is monocistronic, containing information from just one gene. Prokaryotic and eukaryotic tRNAs are also made from longer precursor molecules. If present, an intron is removed by nucleases, and both ends of the molecule are trimmed by ribonucleases. A 3I -CCA sequence is added, and bases at specific positions are modified, producing “unusual” bases.
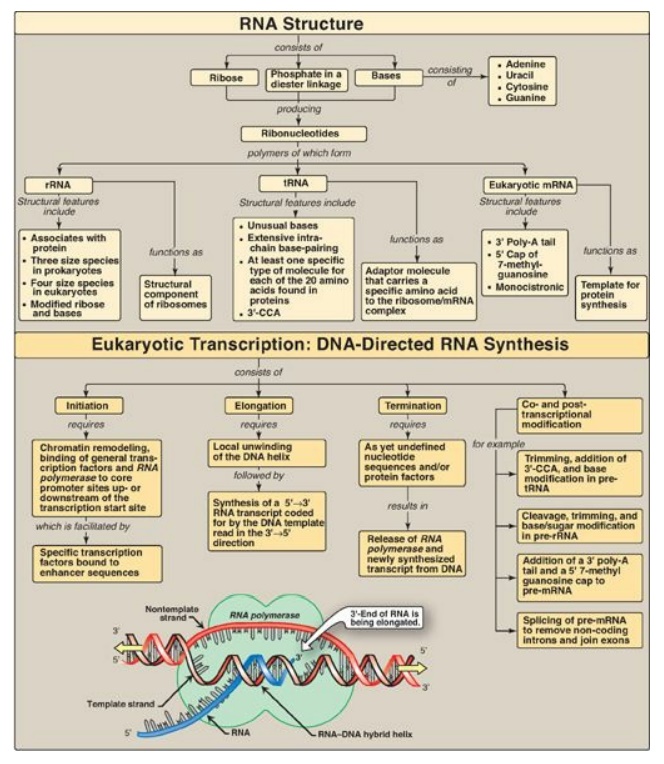
Figure 30.20 Key concept map for RNA structure and synthesis. rRNA = ribosomal RNA; tRNA = transfer RNA; mRNA = messenger RNA.
Study Questions
Choose the ONE correct answer.
30.1 An 8-month-old male with severe anemia is
found to have β-thalassemia. Genetic analysis shows that one of his β-globin
genes has a mutation that creates a new splice acceptor site 19 nucleotides
upstream of the normal splice acceptor site of the first intron. Which of the
following best describes the new messenger RNA molecule that can be produced
from this mutant gene?
A. Exon 1 will be too
short.
B. Exon 1 will be too
long.
C. Exon 2 will be too
short.
D. Exon 2 will be too long.
E. Exon 2 will be
missing.
Correct answer = D. Because the mutation creates an
additional splice acceptor site (the 3I -end) upstream of the normal acceptor
site of intron 1, the 19 nucleotides that are usually found at the 3I -end of
the excised intron 1 lariat can remain behind as part of exon 2. Exon 2 can,
therefore, have these extra 19 nucleotides at its 5I -end. The presence of
these extra nucleotides in the coding region of the mutant messenger RNA (mRNA)
molecule will prevent the ribosome from translating the message into a normal
β-globin protein molecule. Those mRNAs for which the normal splice site is used
to remove the first intron will be normal, and their translation will produce
normal β-globin protein.
30.2 A 4-year-old child who easily tires and has
trouble walking is diagnosed with Duchenne muscular dystrophy, an X-linked
recessive disorder. Genetic analysis shows that the patient’s gene for the
muscle protein dystrophin contains a mutation in its promoter region. Of the
choices listed, which would be the most likely effect of this mutation?
A. Initiation of dystrophin transcription will be
defective.
B. Termination of
dystrophin transcription will be defective.
C. Capping of
dystrophin messenger RNA will be defective.
D. Splicing of
dystrophin messenger RNA will be defective.
E. Tailing of
dystrophin messenger RNA will be defective.
Correct answer = A. Mutations in the promoter
typically prevent formation of the RNA polymerase II transcription complex,
resulting in a decrease in the initiation of messenger RNA (mRNA) synthesis. A
deficiency of dystrophin mRNA will result in a deficiency in the production of
the dystrophin protein. Capping, splicing, and tailing defects are not a
consequence of promoter mutations. They can, however, result in mRNA with
decreased stability (capping and tailing defects), or a mRNA in which too many
or too few introns have been removed (splicing defects).
30.3 A mutation to this sequence in eukaryotic
messenger RNA (RNA) will affect the process by which the 3I -end poly-A tail is
added to the mRNA.
A. AAUAAA
B. CAAT
C. CCA
D. GU…A…AG
E. TATAAA
Correct answer = A. An endonuclease cleaves messenger
RNA just downstream of this polyadenylation signal, creating a new 3I -end to
which the poly A polymerase adds the poly-A tail using ATP as the substrate in
a template-independent process. CAAT and TATAAA are sequences found in
promoters for RNA polymerase II. CCA is added to the 3I -end of transfer RNA by
nucleotidyltransferase. GU…A…AG denotes an intron.
30.4. This protein factor identifies the promoter
of protein-coding genes in eukaryotes.
A. Pribnow box
B. Rho
C. Sigma
D. TFIID
E. U1
Correct answer = D. The general transcription factor,
TFIID, recognizes and binds core promoter elements such as the TATA-like box in
eukaryotic protein-coding genes. These genes are transcribed by RNA polymerase
II. The Pribnow box is a cis-acting element in prokaryotic promoters. Rho is
involved in the termination of prokaryotic transcription. Sigma is the subunit
of prokaryotic RNA polymerase that recognizes and binds the prokaryotic
promoter. U1 is a ribonucleoprotein involved in splicing of eukaryotic
pre-messenger RNA.
30.5 What is the sequence (conventionally written)
of the RNA product of the DNA template sequence, GATCTAC?
Correct answer = 5I -GUAGAUC-3I. The RNA product has a sequence that
is complementary to the template strand, with U replacing T.
Related Topics
