Enumeration of microorganisms
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Fundamental features of microbiology
In a pharmaceutical context there are several situations where it is necessary to measure the number of microbial cells in a culture, sample or specimen:
ENUMERATION OF MICROORGANISMS
In a pharmaceutical context there are several situations where it is necessary to measure the number of microbial cells in a culture, sample or specimen:
• when measuring the levels of microbial contamination in a raw material or manufactured medicine
• when evaluating the effects of an antimicrobial chemical or decontamination process
• when using microorganisms in the manufacture of therapeutic agents
• when assessing the nutrient capability of a growth medium.
In some cases it is necessary to know the total number of microbial cells present, i.e. both living and dead: e.g. in vaccine manufacture dead and living cells may both produce an immune response, and in pyrogen testing both dead and living cells induce fever when injected into the body. However, in many cases it is the number or concentration of living cells that is required. The terminology in microbial counting sometimes causes confusion. A total count is a counting procedure enumerating both living and dead cells, whereas a viable count, which is far more common, records the living cells alone. However, the term total viable count (TVC) is used in most pharmacopoeias and by many regulatory agencies to mean a viable count that records all the different species or types of microorganism that might be present in a sample (e.g. bacteria plus fungi).
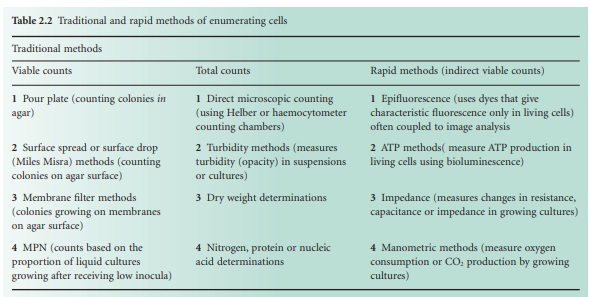
Table 2.2 lists the more common counting methods available. The first three traditional methods of viable counting all operate on the basis that a living cell (or a CFU) will give rise to a visible colony when introduced into or onto the surface of a suitable medium and incubated. Thus, the procedure for pour plating (Figure 2.2A) usually involves the addition of a small volume (typically 1.0 ml) of sample (or a suitable dilution thereof) into molten agar at 45 °C which is then poured into empty sterile Petri dishes. After incubation the resultant colonies are counted and the total is multiplied by the dilution factor (if any) to give the concentration in the original sample. In a surface spread technique (Figure 2.2B) the sample (usually 0.1–0.25 ml) is spread over the surface of agar which has previously been dried to permit absorption of the added liquid. The Miles Misra (surface drop) method (Figure 2.2C) is similar in principle, but several individual drops of culture are allowed to spread over discrete areas of about 1 cm diameter on the agar surface. These procedures are suitable for samples that are expected to contain concentrations exceeding approximately 100 CFU ml −1 so that the number of colonies arising on the plate is sufficiently large to be statistically reliable. If there are no clear indications of the order of magnitude of the concentration in the sample, it is necessary to plate out the sample at each of two, three or more (decimal, i.e. 10-fold) dilutions so as to obtain Petri dishes with conveniently countable numbers of colonies (usually taken to be 30–300 colonies).
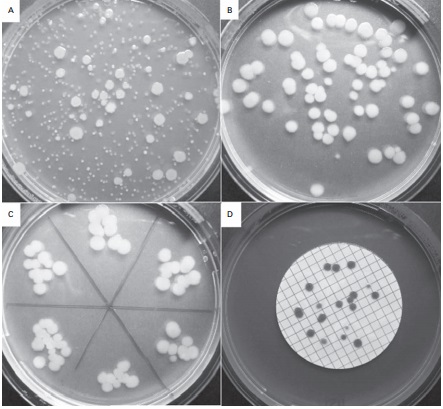
Figure 2.2 Viable counts of bacteria:
(A) Pour plate method using Bacillus subtilis ; the colonies on the surface of
the agar are growing larger than those within the agar due to greater oxygen
availability. (B) Surface spread, and (C) surface drop (Miles Misra) methods
using Bacillus subtilis . (D) Membrane fi ltration method showing Serratia
marcescens colonies growing on a 47 mm diameter membrane.
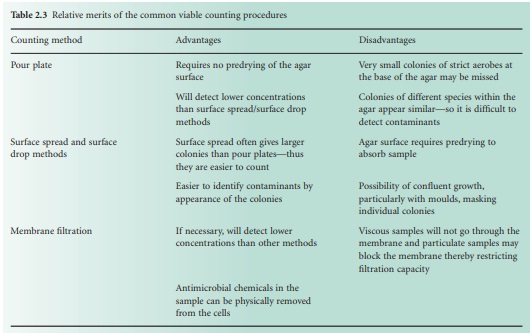
If 30 is accepted as the lowest reliable number to count and a pour plate method uses a 1.0 ml sample, it follows that the procedures described above are unsuitable for any sample that is expected to contain <30 CFU ml −1, e.g. water samples where the count may be 1 CFU ml−1 or less. Here, membrane filter methods are used (Figure 2.2D) in which a large, known volume of sample is passed through the membrane which is placed, without inversion, on the agar surface. Nutrients then diffuse up through the membrane and allow the retained cells to grow into colonies on it just as they would on the agar itself. Some of the relative merits of these procedures are described in Table 2.3.
Most probable number (MPN) counts may be used when the anticipated count is relatively low, i.e. from <1 up to 100 microorganisms ml−1. The procedure involves inoculating multiple tubes of culture medium (usually three or five) with three different volumes of sample, e.g. three tubes each inoculated with 0.1 ml, three with 0.01 ml and three with 0.001 ml. If the concentration in the sample is in the range indicated above, there should be a proportion of the tubes receiving inocula in which no microorganisms are present; these will remain sterile after incubation, while others that received inocula actually containing one or more CFU show signs of growth. The proportions of positive tubes are recorded for each sample volume and the results are compared with standard tables showing the MPN of organisms per ml (or per 100 ml) of original sample. The procedure is more commonly used in the water, food and dairy industries than in the pharmaceutical industry; nevertheless, it is a valid technique described in pharmacopoeias and appropriate for pharmaceutical materials, particularly water. The poor accuracy and precision associated with MPN counts usually means that the method is one of last resort—to be considered only when other counting methods are inappropriate.
Turbidity measurements are the most common means of estimating the total numbers of bacteria present in a sample. Measuring the turbidity using a spectrophotometer or colorimeter and reading the concentration from a calibration plot is a simple means of standardizing cell suspensions for use as inocula in antibiotic assays or other tests of antimicrobial chemicals. Fungi cannot readily be handled in this way because the suspension may not be uniform or may sediment in a spectrophotometer cuvette. Consequently, dry weight determinations on known volumes of culture are an alternative means of estimating fungal biomass. Direct microscopic counting may be an appropriate method for bacteria, yeasts and fungal spores but not for moulds, and indirect measures of biomass like assays of insoluble nitrogen, protein or nucleic acids are possible for all cell types, but rarely used outside the research laboratory.
The traditional methods of viable counting all suffer from the same limitations:
• relatively labour intensive
• not easy to automate
• slow, because they require an incubation period for colonies to develop or liquid cultures to become turbid
• may require relatively large volumes of culture media, many Petri dishes and a lot of incubator space.
For these reasons much interest and investigative effort has been invested in recent years in the use of so-called ‘rapid’ methods of detecting and counting microorganisms. These methods enumerate viable organisms—usually bacteria and yeasts rather than moulds—in a matter of hours and eliminate the 24–48 hour (or longer) incubation periods that are typical of traditional procedures. The rapid methods employ various means of indirect detection of living cells, but the following operating principles are the most common:
• Epi fluorescent techniques use fluorescent dyes that either exhibit different colours in living and dead cells (e.g. acridine orange) or appear colourless outside the cell but become fluorescent when absorbed and subjected to cellular metabolism (e.g. fluorescein diacetate).
• Living cells generate adenosine triphosphate (ATP) that can readily be detected by enzyme assays, e.g. luciferin emits light when exposed to firefly luciferase in the presence of ATP; light emission can be measured and related to bacterial concentration.
• The resistance, capacitance or impedance of a culture medium changes as a result of bacterial or yeast growth and metabolism, and these electrical properties vary in proportion to cell concentration.
• Manometric techniques are appropriate for monitoring the growth of organisms that consume or produce significant quantities of gas during their metabolism, e.g. yeasts or moulds producing carbon dioxide as a result of fermentation.
These methods are fast, readily automated and eliminate the need for numerous Petri dishes and incubators. On the other hand they require expensive equipment, have limitations in terms of detection limits and may be less readily adapted to certain types of sample than traditional methods. Furthermore, there are problems in some cases with reconciling the counts obtained by rapid methods and by traditional means. The newer techniques may detect organisms that are metabolizing but not capable of reproducing to give visible colonies (viable but nonculturable organisms), so may give values many times higher than traditional methods; this has contributed to the caution with which regulatory authorities have accepted the data generated by rapid methods. Nevertheless, they are becoming more widely accepted and are likely to become an integral part of enumeration procedures in pharmaceutical microbiology in the foreseeable future.
Related Topics
