Regulation of Prokaryotic Gene Expression
| Home | | Biochemistry |Chapter: Biochemistry : Regulation of Gene Expression
In prokaryotes such as Escherichia coli (E. coli), regulation of gene expression occurs primarily at the level of transcription and, in general, is mediated by the binding of trans-acting proteins to cis-acting regulatory elements on their single DNA molecule (chromosome).
REGULATION OF PROKARYOTIC GENE EXPRESSION
In prokaryotes such as
Escherichia coli (E. coli), regulation of gene expression occurs primarily at
the level of transcription and, in general, is mediated by the binding of
trans-acting proteins to cis-acting regulatory elements on their single DNA
molecule (chromosome). [Note: Regulating the first step in the expression of a
gene is an efficient approach, insofar as energy is not wasted making unneeded
gene products.] Transcriptional control in prokaryotes can involve the
initiation or premature termination of transcription.
A. Transcription of messenger RNA from bacterial operons
In bacteria, the
structural genes that code for proteins involved in a particular metabolic
pathway are often found sequentially grouped on the chromosome along with the
cis-acting regulatory elements that determine the transcription of these genes.
The transcription product is a single polycistronic messenger RNA (mRNA). The
genes are, thus, coordinately controlled (that is, turned on or off as a unit).
This entire package is referred to as an operon.
B. Role of operators in prokaryotic transcription
Prokaryotic operons
contain an operator, a segment of DNA that regulates the activity of the
structural genes of the operon. If the operator is not bound by a repressor
molecule, RNA polymerase passes over the operator and reaches the
protein-coding genes which it transcribes to mRNA. If a repressor molecule is
bound to the operator, the polymerase is blocked and does not produce mRNA. As
long as the repressor is bound to the operator, no proteins are made. However,
when an inducer molecule is present, it binds to the repressor, causing the
repressor to change shape so that it no longer binds the operator. When this
happens, the RNA polymerase can proceed with transcription. One of the
best-understood examples is the inducible lactose operon of E. coli that
illustrates both positive and negative regulation (Figure 32.4).
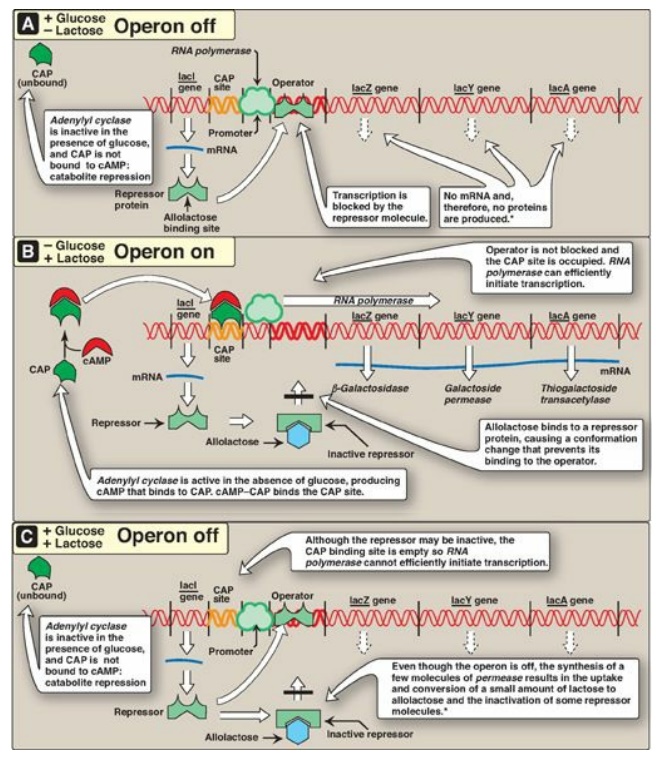
Figure 32.4 The lactose operon of E. coli. *[Note: Even when the operon has been turned off by catabolite repression, the repressor transiently dissociates from the operator at a slow rate, allowing a very low level of expression. The synthesis of a few molecules of permease (and β-galactosidase) allows the organism to respond rapidly should glucose become unavailable.] CAP = catabolite activator protein; cAMP = cyclic adenosine monophosphate; mRNA = messenger RNA.
C. The lactose operon
The lactose (lac) operon contains the genes that code for three proteins involved in the catabolism of the disaccharide lactose: The lacZ gene codes for β-galactosidase, which hydrolyzes lactose to galactose and glucose; the lacY gene codes for a permease, which facilitates the movement of lactose into the cell; and the lacA gene codes for thiogalactoside transacetylase, which acetylates lactose. [Note: The physiologic function of this acetylation is unknown.] All of these proteins are maximally produced only when lactose is available to the cell and glucose is not. [Note: Bacteria use glucose, if available, as a fuel in preference to any other sugar.] The regulatory portion of the operon is upstream of the three structural genes and consists of the promoter region where RNA polymerase binds and two additional sites, the operator (O) site and the CAP site, where regulatory proteins bind. The lacZ, lacY, and lacA genes are expressed only when the O site is empty, and the CAP site is bound by a complex of cyclic adenosine monophosphate ([cAMP]) and the catabolite activator protein (CAP), sometimes called the cAMP regulatory protein (CRP). A regulatory gene, the lacI gene, codes for the repressor protein (a trans-acting factor) that binds to the O site with high affinity. [Note: The lacI gene has its own promoter.]
1. When only glucose is available: In this case, the lac operon is
repressed (turned off). Repression is mediated by the repressor protein binding
via a helix-turn-helix motif (Figure 32.5) to the operator site, which is downstream
of the promoter region (see Figure 32.4A). Binding of the repressor interferes
with the progress of RNA polymerase and blocks transcription of the structural
genes. This is an example of negative regulation.
Figure 32.5 Helix-turn-helix motif of the lac repressor protein.
2. When only lactose is available: In this case, the lac operon is
induced (maximally expressed, or turned on). A small amount of lactose is
converted to an isomer, allolactose. This compound is an inducer that binds to
the repressor protein, changing its conformation so that it can no longer bind
to the operator. In the absence of glucose, adenylyl cyclase is active, and
sufficient quantities of cAMP are made and bind to the CAP protein. The
cAMP–CAP trans-acting complex binds to the CAP site, causing RNA polymerase to
more efficiently initiate transcription at the promoter site (see Figure
32.4B). This is an example of positive regulation. The transcript is a single
polycistronic mRNA molecule that contains three sets of start and stop codons.
Translation of the mRNA produces the three proteins that allow lactose to be
used for energy production by the cell. [Note: In contrast to the inducible
lacZ, lacY, and lacA genes, whose expression is regulated, the lacI gene is
constitutive. Its gene product, the repressor protein, is always made and is active
unless the inducer is present.]
3. When both glucose and lactose are available: In this case, transcription of the lac operon is negligible, even if lactose is present at a high concentration. Adenylyl cyclase is inhibited in the presence of glucose (a process known as catabolite repression) so no cAMP–CAP complex forms, and the CAP site remains empty. RNA polymerase is, therefore, unable to effectively initiate transcription, even though the repressor may not be bound to the operator region. Consequently, the three structural genes of the operon are not expressed (see Figure 32.4C).
D. Tryptophan operon
The tryptophan (trp)
operon contains five structural genes that code for enzymes required for the
synthesis of the amino acid, tryptophan. As with the lac operon, the trp operon
is subject to negative control. However, for the repressible trp operon,
negative control includes Trp itself binding to a repressor protein and
facilitating the binding of the repressor to the operator: Trp is a
corepressor. Because repression by Trp is not always complete, unlike the lac
operon, the trp operon is also regulated by a process known as attenuation.
With attenuation, transcription is initiated but is terminated well before
completion (Figure 32.6). If Trp is plentiful, transcription initiation that
escaped repression by Trp is attenuated (stopped) by the formation at the 5I
-end of the mRNA of a hairpin (stem–loop) structure like that seen in
rho-independent termination. [Note: Transcription and translation are
temporally linked in prokaryotes, and, therefore, attenuation also results in
the formation of a truncated, nonfunctional peptide product that is rapidly
degraded.] If Trp becomes scarce, the operon is expressed. The 5I -end of the
mRNA contains two adjacent codons for Trp. The lack of Trp causes ribosomes to
stall at these codons, covering regions of the mRNA required for formation of
the attenuation hairpin. This prevents attenuation and allows transcription to
continue.
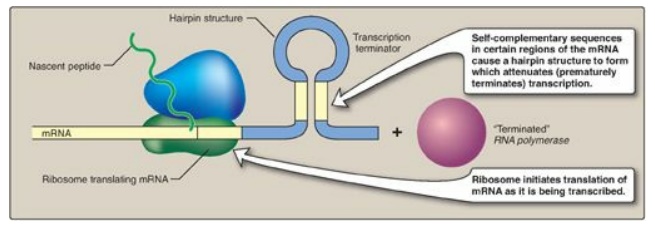
Figure 32.6 Attenuation of
transcription of the trp operon when tryptophan is plentiful. mRNA = messenger
RNA.
Transcriptional attenuation can occur in
prokaryotes because translation of an mRNA begins before its synthesis is
complete. This does not occur in eukaryotes because the presence of a
membrane-bound nucleus spatially and temporally separates transcription and
translation.
E. Coordination of transcription and translation in prokaryotes
Whereas transcriptional
regulation of mRNA production is primary in bacteria, regulation at the level
of ribosomal RNA (rRNA) and protein synthesis also occurs and plays important
roles in the microbe’s ability to adapt to environmental stress.
1. Stringent response: E. coli has seven operons that
synthesize the rRNA needed for ribosome assembly, and each is regulated in
response to changes in environmental conditions. Regulation in response to
amino acid starvation is known as the stringent response. The binding of an
uncharged transfer RNA (tRNA) to the A site of a ribosome triggers a series of
events that leads to the production of a polyphosphorylated guanosine, ppGpp.
The synthesis of this unusual derivative of guanosine diphosphate (GDP) is catalyzed
by stringent factor (RelA), an enzyme physically associated with ribosomes.
Elevated levels of ppGpp result in inhibition of rRNA synthesis (Figure 32.7).
[Note: In addition to rRNA, tRNA synthesis and some mRNA synthesis (for
example, for ribosomal proteins) are also inhibited. However, synthesis of
mRNAs for enzymes required for amino acid biosynthesis is not inhibited. ppGpp
appears to alter promoter selection through use of different sigma-factors for
RNA polymerase.]
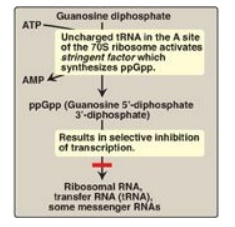
Figure 32.7 Regulation of
transcription by the stringent response to amino acid starvation. S = Svedberg
unit.
2. Regulatory ribosomal proteins: Operons for ribosomal proteins
(r-proteins) can be inhibited by an excess of their own protein products. For
each operon, one specific r-protein functions in the repression of translation
of the polycistronic mRNA from that operon (Figure 32.8). The r-protein does so
by binding to the Shine-Dalgarno (SD) sequence located on the mRNA just
upstream of the first initiating AUG codon and acting as a physical impediment
to the binding of the small ribosomal subunit to the SD sequence. One r-protein
thus inhibits synthesis of all the r-proteins of the operon. This same
r-protein also binds to rRNA and with a higher affinity than for mRNA. If the
concentration of rRNA falls, the r-protein then is available to bind its own
mRNA and inhibit its translation. This coordinated regulation keeps the
synthesis of r-proteins in balance with the transcription of rRNA, so that each
is present in appropriate amounts for the formation of ribosomes.
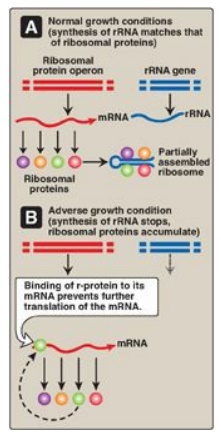
Figure 32.8 Regulation of
translation by an excess of ribosomal proteins. mRNA = messenger RNA; rRNA =
ribosomal RNA; r-protein = ribosomal protein.
Related Topics
