DNA Based Technologies - Methods of Detection of Parasites
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Protozoa
Nucleic acids, DNA or RNA, are found in all types of microorganism and the sequences of these molecules can be used to help in the identification of species and individuals.
DNA BASED TECHNOLOGIES
Nucleic acids, DNA or
RNA, are found in all types of microorganism and the sequences of these
molecules can be used to help in the identification of species and individuals.
The robust nature of DNA has allowed researchers to use these sequence in a
variety applications including forensics, archaeology (sequences from animal
tissues over 100 years old have routinely been determined) and epidemiology.
The advent of highly sensitive DNA amplification technologies such as the
polymerase chain reaction (PCR) has allowed the development of molecular tools
for the identification and detection of parasites. Initial work on these
techniques focused on their ability to help define species and many of these
studies utilized the ribosomal DNA genes and were very successful. Using these
genes it was possible to discriminate many genera and species, but the genes
lacked the level of sequence variation required for separation below the level
of species (important for some pathogenic organisms and for epidemiological
studies). In addition ribosomal DNA sequencing was also not always appropriate
for identifying species that are highly diverse. Other DNA markers that have
been used included genes that encode for metabolic enzymes and structural
proteins. For example Cryptosporidium
can be speciated by using a combination of target genes including the Cryptosporidium oocyst wall protein
(COWP), heat shock protein (hsp70), dihydrofolate reductase (DHFR) and 18S ribosomal
DNA. Complete genomes are now available for the major protozoan parasites and
this has helped in the development of improved methods for species
identification. These genomes can be accessed at http://eupathdb.org/eupathdb/
and this website links to many others resources including the National Centre for
Biotechnology Information (http:// www.ncbi.nlm.nih.gov/). Other DNAbased
techniques are now available such as the multiplex PCR (amplification of more
than one gene at a time) and quantitative real time PCR (qPCR), and these have
improved specificity and sensitivity in detection and increased the usefulness
of DNA technology for the detection of parasites in environmental as well as
clinical samples. It is also possible to assess the viability of protozoan
parasites using reverse transcription PCR (RTPCR). This is due to the fact that
mRNAs degrade quickly once parasites are killed. However, it should be noted
that increasing evidence exists to show that mRNA can survive in some cells for
up to 24 hours
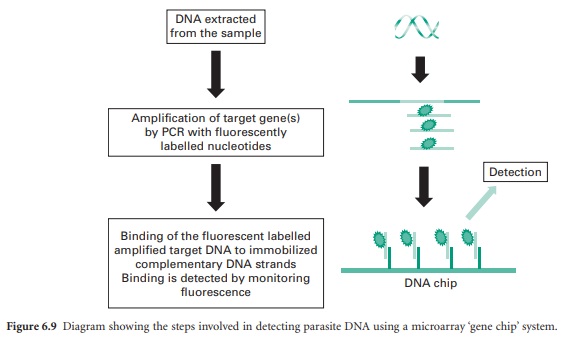
A recent method, DNA
microarray, is a fast developing technology. This is a ‘chip’ based system that
measures the binding of target DNA sequences (parasite) from samples
(fluorescently labelled using PCR) to complementary sequences bound to the chip
surface. The binding of parasite sequence DNA and the amount that is bound can
be detected using fluorescence (Figure 6.9). In theory this technique allows
the detection of multiple parasite species at the same time in one sample.
Related Topics
