Genetic Methods of Classifying Microbes
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Characterization, Classification and Taxonomy of Microbes
There are three most prominent ‘genetic methods’ that are invariably employed for the methodi-cal arrangement of microbes based upon various taxonomic groups (i.e., Taxa), namely: (i) Genetic relatedness (ii) The intuitive method, and (iii) Numerical taxonomy.
Genetic
Methods of Classifying Microbes
There are
three most prominent ‘genetic methods’ that are invariably
employed for the methodi-cal arrangement of microbes based upon various
taxonomic groups (i.e., Taxa),
namely:
(i) Genetic
relatedness
(ii) The
intuitive method, and
(iii) Numerical
taxonomy.
The
aforesaid ‘genetic methods’ shall now
be treated separately in the sections that follows.
1. Genetic Relatedness
It is
regarded to be one of the most trustworthy and dependable method of
classification based solely upon the critical extent of genetic relatedness occurring between different organisms. In
addition this particular method is considered not only to be the utmost
objective of all other techniques based upon the greatest extent pertaining to
the fundamental aspect of organisms, but also their inherent he-reditary
material (deoxyribonucleic acid, DNA).
It is,
however, pertinent to state here that in actual practice the genetic relatedness may also be
estimated by precisely measuring the degree of hybridization taking place either between denatured DNA molecules
or between single stranded DNA and RNA species. The extent of homology* is as-sayed by strategically
mixing two different, types of ‘single-stranded DNA’ or ‘single-stranded DNA with RNA’ under highly specific and
suitable experimental parameters; and subsequently, measuring accurately the degree to which they
are actually and intimately associated to give rise to the formation of the
desired ‘double-stranded structures’
ultimately. The aforesaid aims and objectives may be accom-plished most
precisely and conveniently by rendering either the DNA or RNA radioactive and
measur-ing the radio activities by the help of Scintillation Counter or Geiger-Müller
Counter.
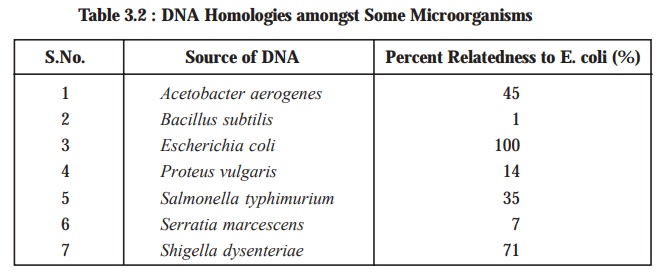
Table
3.2, shows the extent of genetic
relatedness of different microbes as assayed by the ensu-ing DNA-RNA hybridization. Nevertheless, it
has been duly demonstrated and proved that the genetic relatedness can be
estimated accurately by DNA-RNA
hybridization; however, the DNA-DNA
hybridi-zation affords the most precise results, provided adequate
precautions are duly taken to ascertain and
ensure that the prevailing hybridization
between the two strands is perfectly uniform.
2. The Intuitive Method
Various ‘microbiologists’ who have acquired
enormous strength of knowledge, wisdom, and hands-on experience in the
expanding field of ‘microbiology’
may at a particular material time vehe-mently decide and pronounce their
ultimate verdict whether the microorganisms represent one or more species or
genera. The most predominant and utterly important disadvantage of this particular method being that the
characteristic features of an organism which may appear to be critical and
vital to one researcher may not seem to be important to the same extent to
another, and altogether different taxono-mists would ultimately decide on
something quite different categorization at the end. Nevertheless, there are
certain ‘classification schemes’
that are exclusively based upon the intuitive
method and definitively proved to be immensely beneficial and useful in microbiology.
3. Numerical Taxonomy
The
survey of literatures have amply proved that in the Nineteenth Century, microbes were categorically grouped
strictly in proportion to their evolutionary affinities. Consequently, the
systematic and methodical segregation and arrangement of microorganisms into
the various organized groups was entirely on the specialized foundation of
inherited and stable structural and physiological characteristic features. This
arrangement is termed as the ‘Natural
Classification’ or the ‘Phylogenetic
Classifiction’.
Interestingly,
this particular modus operandi for
the classification of microorganisms has now almost turned out to be absolutely
redundant, and hence abandoned outright quite in favour of a rather more
realistic empirical approach based exclusively on ‘precise quantification’ pertaining to close similarities and distinct
dissimilarities prevailing amongst the various microbes. Michael Adanson was the first and
foremost microbiologist who unequivocally suggested this magnanimous approach,
which was termed as Adansonian Taxonomy
or Numerical Taxonomy.
Salient Features: The
various salient features of the
Numerical Taxonomy (or Adansonian
Taxonomy) are as enumerated below:
(1) The
fundamental basis of Numerical Taxonomy
is the critical assumption, that in the event when each phenotypic character is
assigned even and equal weightage, it must be viable and feasible to express
numerically the explicit taxonomic
distances existing between microor-ganisms, with regard to the number of actual characters which are shared in
comparison to the total number of
characters being examined ultimately. The importance of the Numerical Taxonomy is largely
influenced by the number of characters being investigated. Therefore, it would be absolutely necessary to
accomplish precisely an extremely high degree of signifi-cance—one should
examine an equally large number of characters.
(2) Similarity Coefficient and Matching
Coefficient: The determination of the similarity co efficient as well as the matching coefficient of any two
microbial strains, as characterized with regard to several character
variants viz., a, b, c, d etc., may
be determined as stated under:
Number of
characters + ve in both strains = a
Number of
characters + ve in ‘strain-1’ and – ve in ‘Strain-2’ = b
Number of
characters, – ve in ‘Strain-1’ and + ve in ‘Strain-2’ = c
Number of
characters – ve in both strain = d
Similarity
coefficient [Sj] = a / ( a + b + c )
Matching
coefficient [Ss] = a + b / ( a + b + c + d )
Based on
the results obtained from different experimental designs, it has been observed
that the similarity coefficient does
not take into consideration the characters that are ‘negative’ for both organ-isms; whereas, the matching coefficient essentially includes both positive and
negative characters.
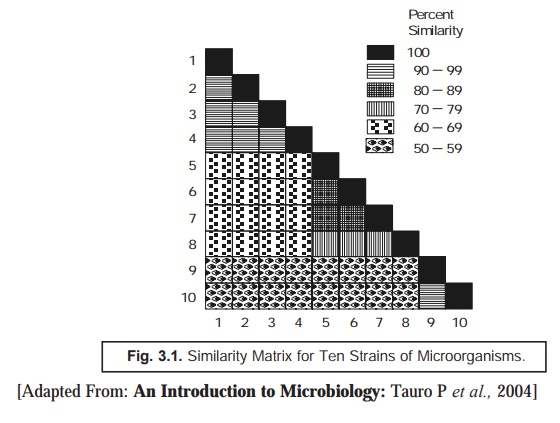
Similarity Matrix: The ‘data’ thus generated are carefully
arranged in a ‘similarity matrix’ only
after having estimated the similarity
coefficient and the matching
coefficient for almost all microor-ganisms under investigation duly and
pair-wise, as depicted in Fig. 3.1 below. Subsequently, all these matrices may
be systematically recorded to bring together the identical and similar strains
very much close to one another.
In actual
practice, such data are duly incorporated and transposed to a ‘dandogram’* as illus-trated in Fig.
3.2 under, that forms the fundamental basis for establishing the most probable
taxonomic
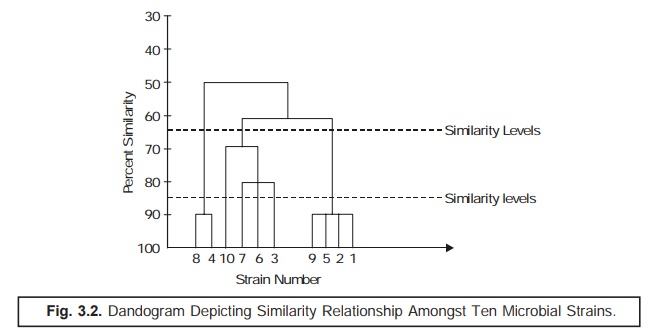
The ‘dotted line’ as indicated in
(Fig. 3.2) a dandogram evidently shows ‘similarity
levels’ that might be intimately
taken into consideration for recognizing
two different taxonomic ranks, for instance: a genus and a species.
The ‘Numerical Taxonomy’ or ‘Adansonian Approach’ was thought and
believed to be quite impractical and cumbersome in actual operation on account
of the reasonably copious volume and mag-nitude of the ensuing numerical
calculations involved directly. Importantly, this particular aspect has now
almost been eliminated completely by the advent of most sophisticated ‘computers’ that may be programmed
appropriately for the computation of the data, and ultimately, arrive at the degree of simi-larity with great ease,
simplicity, and precision. It is, however, pertinent to point out at this
juncture that though the ensuing ‘Numerical Taxonomy’ fails to throw any
light with specific reference to the pre-vailing genetic relationship, yet it
amply gives rise to a fairly stable fundamental basis for the articu-lated categorization of the
taxonomic distribution and groupings.
Limitations of Numerical Taxonomy:
The
various limitations of numerical
taxonomy are as enumerated
under:
(1) It is
useful to classify strains within a larger group which usually shares the
prominent characteristic features in common.
(2) The
conventional classification of organisms solely depends on the observations and
knowledge of the individual taxonomist in particular to determine the ensuing
matching similarities existing between the bacterial strains; whereas, numerical taxonomy exclusively depends
upon the mathematical figures plotted on paper.
(3) The actual usage of several tests reveals a good number of phenotypes, thereby more genes are being screened; and, therefore, no organism shall ever be missed in doing so.
(4) One
major limitation of the numerical
analysis is that in some instances, a specific strain may be grouped with a
group of strains in accordance to the majority of identical characteristic
features, but certainly not to all the prevailing characters. However,
simultaneously the particu-lar strain may possess a very low ebb of similarity
with certain other members of the cluster.
(5) The
exact location of the taxon is not
yet decided, and hence cannot be grouped or related to any particular taxonomic
group, for instance : genes or species.
(6) Evidently,
in the numerical analysis, the
definition of a species is not
acceptable as yet, whereas some surveys do ascertain that a 65% single-linkage cluster distincly
provides a 75% approximate idea of the specific species.
Related Topics
