Chromosome Function and Replication - Basis for the Selective Inhibition of Chromosome Replication and Function
| Home | | Pharmaceutical Microbiology | | Pharmaceutical Microbiology |Chapter: Pharmaceutical Microbiology : Mechanisms of action of antibiotics and synthetic anti-infective agents
As with protein synthesis, the mechanisms of chromosome replication and function are essentially the same in prokaryotes and eukaryotes. There are, however, important differences in the detailed functioning and properties of the enzymes involved and these differences are exploited by a number of agents as the basis of selective inhibition.
CHROMOSOME FUNCTION AND
REPLICATION
BASIS FOR THE SELECTIVE INHIBITION OF CHROMOSOME REPLICATION AND FUNCTION
As with protein synthesis, the mechanisms of chromosome replication and function are essentially the same in prokaryotes
and eukaryotes. There are, however, important differences in the detailed functioning and properties of the enzymes involved and these differences are exploited by a number
of agents as the basis
of selective inhibition. The microbial chromosome is large in comparison with the cell
that contains it (approximately 500 times the length
of E. coli). It is therefore wound into a compact, supercoiled form inside the cell. During
replication the circular double
helix must be unwound to allow the DNA polymerase enzymes
to synthesize new complementary strands. The shape of the chromosome is
manipulated by the cell by the formation of regions of supercoiling.
Positive supercoiling (coiling in the same
sense as the turns
of the double helix) makes
the chromosome more compact. Negative
supercoiling (generated by twisting the chromosome in the opposite
sense to the helix) produces localized strand
separation which is required both for
replication and transcription. In a bacterium such as E. coli four different topoisomerase
enzymes are responsible for maintaining the shape of DNA
during cell division. They act by cutting one or both of
the DNA strands; they
remove and generate supercoiling, then reseal
the strands. Their
activity is essential for the microbial cell to relieve
the complex tangling
of the chromosome (both
knotting and chain
link formation) which results
from progression of the replication fork
around the circular
chromosome. Type I topoisomerases cut one strand
of DNA and pass
the other strand
through the gap before resealing. Type II enzymes cut both strands and pass another
double helical section
of the DNA through the gap before resealing. In E. coli topoisomerases I and III are both
type I enzymes
while topoisomerases II and IV are type II enzymes.
Topoisomerase II (also known as DNA gyrase) and topoisomerase IV are essential enzymes which are inhibited by the fluoroquinolone group of antimicrobials. Topoisomerase II is responsible for introducing negative supercoils into DNA and for
relieving torsional stress, which accumulates ahead of
sites of transcription and replication. Topoiosomerase IV provides a potent
decatenating (unlinking) activity that removes
links and knots generated behind the replication fork.
The basic sequence of events for microbial chromosome replication is described below.
a)
Synthesis Of Precursors
Purines, pyrimidines and their nucleosides and
nucleoside triphosphates are synthesized in the cytoplasm. At this stage the antifolate drugs (sulphonamides and dihydrofolate reductase
inhibitors) act by interfering with the
synthesis and recycling of the cofactor
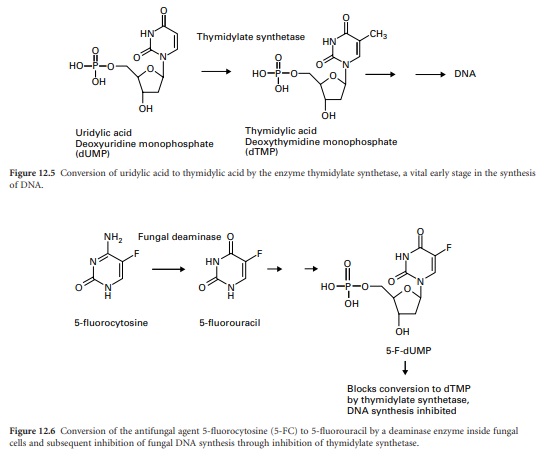
dihydrofolic acid (DHF). Thymidylic acid (2-deoxythymidine monophosphate, dTMP) is an essential nucleotide precursor of DNA synthesis. It is produced
by the enzyme thymidylate
synthetase by transfer
of a methyl group from
tetrahydrofolic acid (THF) to the uracil base on uridylic acid (2-deoxyuridine monophosphate, dUMP) (Figure 12.5). THF is converted to DHF in this process and must be reverted to THF by the
enzyme dihydrofolate reductase (DHFR) before the cycle
can be repeated. By inhibiting DHFR, the antifolates effectively block the production of dTMP and hence DNA synthesis.
The antifungal agent 5-fluorocytosine also
interferes with these early
stages of DNA synthesis. Through conversion to the nucleoside triphosphate it subsequently blocks thymidylic acid production through
inhibition of the enzyme thymidylate synthetase (Figure 12.6).
The antiviral
nucleosides aciclovir and ganciclovir are also converted to their respective nucleoside triphosphates in the cytoplasm of infected
cells. They proceed to inhibit viral
DNA replication either by inhibition of the DNA polymerase or by incorporation into DNA with subsequent termination of chain extension. Finally, the anti-HIV
drug AZT acts in an analogous
manner, being converted to the corresponding triphosphate and inhibiting viral RNA synthesis by the HIV reverse transcriptase.
b)
Unwinding Of The Chromosome
As described in section 4.1,
the DNA double
helix must unwind to allow
access of the polymerase enzymes to
produce two new strands of DNA. This is facilitated by topoisomerase II (DNA gyrase) which
is the target of the fluoroquinolones. Some agents interfere with the unwinding of the chromosome by physical obstruction. These include the acridine dyes, of which
the topical antiseptic proflavine is the most familiar,
and the antimalarial acridine mepacrine. They prevent strand separation by insertion (intercalation) between base pairs
from each strand, but exhibit very poor selective
toxicity.
c)
Replication Of DNA Strands
The unwound DNA strands are kept unfolded during
replication by binding
a protein called
Albert’s protein. A series of enzymes
produce new strands
of DNA using each of the separated strands
as templates. One strand is produced continuously. The other
is produced in a series of short strands called Okazaki fragments
that are joined by a DNA ligase.
The entire process
is carefully regulated, with proofreading stages to check that each nucleotide is correctly
incorporated as specified by the template
sequence. There are no therapeutic agents yet known which interfere directly with the DNA polymerases.
d)
Transcription
The process
of transcription, the copying of a single strand of mRNA sequence using
one strand of the chromosome as a template, is carried out
by RNA polymerase. This is a complex of four proteins
(2 α, 1 β and 1 β′ subunits) which make up the core enzyme.
Another small protein, the σ factor, joins
the core enzyme,
which binds to the promoter
region of the DNA preceding the gene
that is to be transcribed. The correct positioning and orientation of the polymerase is obtained by recognition
of specific marker sites on the DNA at positions −10 and −35 nucleotide bases before
the initiation site for transcription. The σ factor
is responsible for recognition of the initiation signal
for transcription and the core enzyme
possesses the activity
to join the nucleotides in the
sequence specified by the gene. Mammalian genes possess an analogous RNA polymerase but there are sufficient
differences in structure to permit selective inhibition of the
microbial enzyme by the semisynthetic rifamycin antibiotics rifampicin and rifabutin.
Related Topics
