Eukaryotic DNA Replication
| Home | | Biochemistry |Chapter: Biochemistry : DNA Structure, Replication, and Repair
The process of eukaryotic DNA replication closely follows that of prokaryotic DNA synthesis.
EUKARYOTIC DNA REPLICATION
The process of
eukaryotic DNA replication closely follows that of prokaryotic DNA synthesis.
Some differences, such as the multiple origins of replication in eukaryotic
cells versus single origins of replication in prokaryotes, have already been
noted. Eukaryotic single-stranded DNA-binding proteins and ATP-dependent DNA
helicases have been identified, whose functions are analogous to those of the
prokaryotic enzymes previously discussed. In contrast, RNA primers are removed
by RNase H and flap endonuclease-1 (FEN1) rather than by a DNA polymerase
(Figure 29.21).
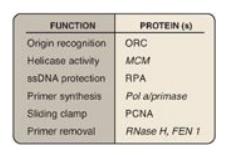
Figure 29.21 Proteins and their function in eukaryotic replication. ORC = origin recognition complex, MCM = minichromosome maintenance (complex), RPA = replication protein A, PCNA = proliferating cell nuclear antigen.
A. Eukaryotic cell cycle
The events surrounding eukaryotic DNA replication and cell division (mitosis) are coordinated to produce the cell cycle (Figure 29.22). The period preceding replication is called the G1 phase (Gap 1). DNA replication occurs during the S (synthesis) phase. Following DNA synthesis, there is another period (G2 phase, or Gap2) before mitosis (M). Cells that have stopped dividing, such as mature T lymphocytes, are said to have gone out of the cell cycle into the G0 phase. Such quiescent cells can be stimulated to reenter the G1 phase to resume division. [Note: The cell cycle is controlled at a series of “checkpoints” that prevent entry into the next phase of the cycle until the preceding phase has been completed. Two key classes of proteins that control the progress of a cell through the cell cycle are the cyclins and cyclin-dependent kinases (Cdks).]
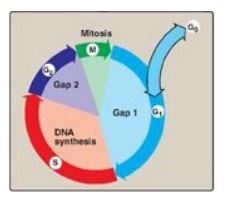
Figure 29.22 The eukaryotic cell cycle. [Note: Cells can leave the cell cycle and enter a reversible quiescent state called G0.]
B. Eukaryotic DNA polymerases
At least five
high-fidelity eukaryotic DNA polymerases (pol) have been identified and
categorized on the basis of molecular weight, cellular location, sensitivity to
inhibitors, and the templates or substrates on which they act. They are
designated by Greek letters rather than by Roman numerals (Figure 29.23).
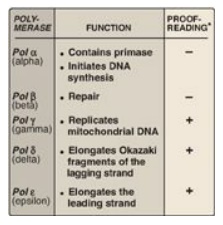
Figure 29.23 Activities of eukaryotic DNA polymerase (pol) *3 →5 exonuclease activity.
1. Pol α: Pol α is a multisubunit enzyme. One subunit has
primase activity, which initiates strand synthesis on the leading strand and at
the beginning of each Okazaki fragment on the lagging strand. The primase
subunit synthesizes a short RNA primer that is extended by the 5 I →3I polymerase activity of pol α, generating a short piece of DNA.
[Note: Pol α is also referred to as pol α/primase.]
2. Pol ε and pol d: Pol ε is recruited to complete DNA
synthesis on the leading strand, whereas pol d elongates the Okazaki fragments
of the lagging strand, each using 3I →5I exonuclease activity to proofread the
newly synthesized DNA. [Note: DNA pol ε associates with proliferating cell
nuclear antigen (PCNA), a protein that serves as a sliding DNA clamp in much
the same way the β subunit of DNA pol III does in E. coli, thus ensuring high
processivity.]
3. Pol β and pol γ: Pol β is involved in “gap filling” in DNA repair (see below). Pol γ replicates mitochondrial DNA.
C. Telomeres
Telomeres are complexes
of noncoding DNA plus proteins (collectively known as shelterin) located at the
ends of linear chromosomes. They maintain the structural integrity of the
chromosome, preventing attack by nucleases, and allow repair systems to
distinguish a true end from a break in dsDNA. In humans, telomeric DNA consists
of several thousand tandem repeats of a noncoding hexameric sequence, AGGGTT,
base-paired to a complementary region of Cs and As. The GT-rich strand is
longer than its CA complement, leaving ssDNA a few hundred nucleotides in
length at the 3I -end. The
single-stranded region is thought to fold back on itself, forming a loop
structure that is stabilized by protein.
1. Telomere shortening: Eukaryotic cells face a special
problem in replicating the ends of their linear DNA molecules. Following removal
of the RNA primer from the extreme 5I
-end of the lagging strand, there is no way to fill in the remaining gap with
DNA. Consequently, in most normal human somatic cells, telomeres shorten with
each successive cell division. Once telomeres are shortened beyond some
critical length, the cell is no longer able to divide and is said to be
senescent. In germ cells and other stem cells, as well as in cancer cells,
telomeres do not shorten and the cells do not senesce. This is a result of the
presence of a ribonucleoprotein, telomerase, which maintains telomeric length
in these cells.
2. Telomerase: This complex contains a protein (Tert) that acts as
a reverse transcriptase and a short piece of RNA (Terc) that acts as a
template. The CA-rich RNA template base-pairs with the GT-rich, single-stranded
3I -end of telomeric DNA (Figure
29.24). The reverse transcriptase uses the RNA template to synthesize DNA in
the usual 5 I →3I direction, extending the already longer 3I -end. Telomerase then translocates to the newly synthesized end,
and the process is repeated. Once the GT-rich strand has been lengthened,
primase activity of DNA pol α can use it as a template to synthesize an RNA
primer. The RNA primer is extended by DNA pol α, and then removed.
Telomeres may be viewed as mitotic clocks in that
their length in most cells is inversely related to the number of times the
cells have divided. The study of telomeres provides insight into the biology of
aging and cancer.
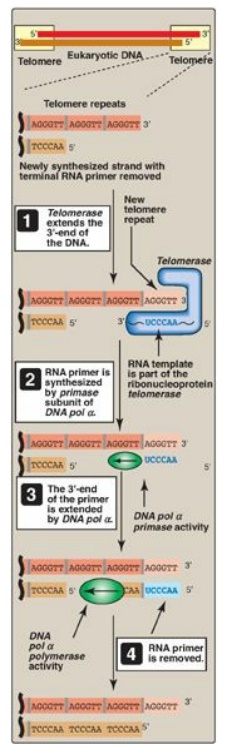
Figure 29.24 Mechanism of
action of telomerase. T= thymine; A = adenine; C = cytosine; G = guanine; pol =
polymerase.
D. Reverse transcriptases
As seen with
telomerase, reverse transcriptases are RNA-directed DNA polymerases. A reverse
transcriptase is involved in the replication of retroviruses, such as human
immunodeficiency virus (HIV). These viruses carry their genome in the form of
ssRNA molecules. Following infection of a host cell, the viral enzyme reverse
transcriptase uses the viral RNA as a template for the 5I →3I synthesis of viral
DNA, which then becomes integrated into host chromosomes. Reverse transcriptase
activity is also seen with transposons, DNA elements that can move about the
genome. In eukaryotes, such elements are transcribed to RNA, the RNA is used as
a template for DNA synthesis by a reverse transcriptase encoded by the
transposon, and the DNA is randomly inserted into the genome. [Note:
Transposons that involve an RNA intermediate are called retrotransposons or
retroposons.]
E. Inhibition of DNA synthesis by nucleoside analogs
DNA chain growth can be
blocked by the incorporation of certain nucleoside analogs that have been
modified on the sugar portion (Figure 29.25). For example, removal of the
hydroxyl group from the 3I -carbon
of the deoxyribose ring as in 2I ,3I - dideoxyinosine ([ddI] also known as
didanosine), or conversion of the deoxyribose to another sugar, such as
arabinose, prevents further chain elongation. By blocking DNA replication,
these compounds slow the division of rapidly growing cells and viruses.
Cytosine arabinoside (cytarabine, or araC) has been used in anticancer
chemotherapy, whereas adenine arabinoside (vidarabine, or araA) is an antiviral
agent. Substitution on the sugar moiety, as seen in zidovudine (AZT, ZDV), also
terminates DNA chain elongation. [Note: These drugs are generally supplied as
nucleosides, which are then converted to the active nucleotides by cellular
kinases.
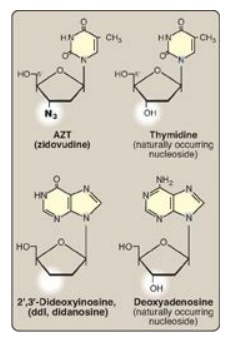
Figure 29.25 Examples of
nucleoside analogs that lack a 3I -hydroxyl group.
[Note: ddI is converted to its active form (ddATP).]
Related Topics
