Structure of DNA
| Home | | Biochemistry |Chapter: Biochemistry : DNA Structure, Replication, and Repair
DNA is a polymer of deoxyribonucleoside monophosphates (dNMPs) covalently linked by 3→5–phosphodiester bonds.
STRUCTURE OF DNA
DNA is a polymer of
deoxyribonucleoside monophosphates (dNMPs) covalently linked by
3→5–phosphodiester bonds. With the exception of a few viruses that contain
single-stranded (ss) DNA, DNA exists as a double-stranded (ds) molecule, in
which the two strands wind around each other, forming a double helix. [Note:
The sequence of the linked dNMPs is primary structure, whereas the double helix
is secondary structure.] In eukaryotic cells, DNA is found associated with
various types of proteins (known collectively as nucleoprotein) present in the
nucleus, whereas in prokaryotes, the protein– DNA complex is present in a
nonmembrane-bound region known as the nucleoid.
A. 3 →5 -Phosphodiester bonds
Phosphodiester bonds
join the 3-hydroxyl group of the deoxypentose of one nucleotide to the
5-hydroxyl group of the deoxypentose of an adjacent nucleotide through a
phosphoryl group (Figure 29.2). The resulting long, unbranched chain has
polarity, with both a 5-end (the end with the free phosphate) and a 3-end (the
end with the free hydroxyl) that are not attached to other nucleotides. The
bases located along the resulting deoxyribose–phosphate backbone are, by
convention, always written in sequence from the 5-end of the chain to the
3-end. For example, the sequence of bases in the DNA shown in Figure 29.2D (5I -TACG-3I) is read “thymine, adenine, cytosine, guanine.” Phosphodiester
linkages between nucleotides can be hydrolyzed enzymatically by a family of
nucleases: deoxyribonucleases for DNA and ribonucleases for RNA, or cleaved
hydrolytically by chemicals. [Note: Only RNA is cleaved by alkali.]
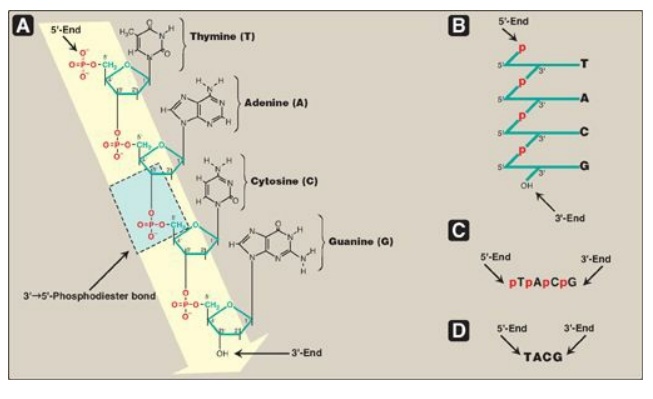
Figure 29.2 A. DNA chain with
the nucleotide sequence shown written in the 5I
→3 I direction. A 3
I →5 I -phosphodiester
bond is shown highlighted in the blue box, and the deoxyribose-phosphate
backbone is shaded in yellow. B. The DNA chain written in a more stylized form,
emphasizing the deoxyribose-phosphate backbone. C. A simpler representation of
the nucleotide sequence. D. The simplest (and most common) representation, with
the abbreviations for the bases written in the conventional 5
I →3 I direction.
B. Double helix
In the double helix,
the two chains are coiled around a common axis called the helical axis. The
chains are paired in an antiparallel manner (that is, the 5 I -end of one strand is paired with the 3 I -end of the other strand) as shown in Figure 29.3. In the DNA
helix, the hydrophilic deoxyribose–phosphate backbone of each chain is on the
outside of the molecule, whereas the hydrophobic bases are stacked inside. The
overall structure resembles a twisted ladder. The spatial relationship between
the two strands in the helix creates a major (wide) groove and a minor (narrow)
groove. These grooves provide access for the binding of regulatory proteins to
their specific recognition sequences along the DNA chain. [Note: Certain anticancer
drugs, such as dactinomycin (actinomycin D), exert their cytotoxic effect by
intercalating into the narrow groove of the DNA double helix, thereby
interfering with DNA (and RNA) synthesis.]
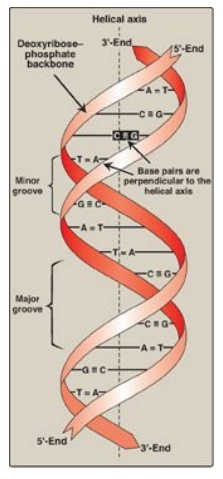
Figure 29.3 DNA double helix, illustrating some of its major structural features.
1. Base-pairing: The bases of one strand of DNA are paired with the
bases of the second strand, so that an adenine (A) is always paired with a
thymine (T) and a cytosine (C) is always paired with a guanine (G). [Note: The
base pairs are perpendicular to the helical axis (see Figure 29.3).] Therefore,
one polynucleotide chain of the DNA double helix is always the complement of
the other. Given the sequence of bases on one chain, the sequence of bases on
the complementary chain can be determined (Figure 29.4). [Note: The specific
base-pairing in DNA leads to the Chargaff rule, which states that in any sample
of dsDNA, the amount of A equals the amount of T, the amount of G equals the
amount of C, and the total amount of purines equals the total amount of
pyrimidines.] The base pairs are held together by hydrogen bonds: two between A
and T and three between G and C (Figure 29.5). These hydrogen bonds, plus the
hydrophobic interactions between the stacked bases, stabilize the structure of
the double helix.
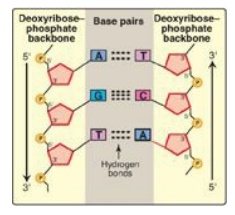
Figure 29.4 Two complementary DNA sequences. T= thymine; A = adenine; C = cytosine; G = guanine.
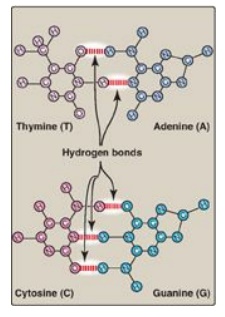
Figure 29.5 Hydrogen bonds between complementary bases.
2. Separation of the two DNA strands in the double
helix: The
two strands of the double helix separate when hydrogen bonds between the paired
bases are disrupted. Disruption can occur in the laboratory if the pH of the
DNA solution is altered so that the nucleotide bases ionize, or if the solution
is heated. [Note: Phosphodiester bonds are not broken by such treatment.] When
DNA is heated, the temperature at which one half of the helical structure is
lost is defined as the melting temperature (Tm). The loss of helical structure
in DNA, called denaturation, can be monitored by measuring its absorbance at
260 nm. [Note: ssDNA has a higher relative absorbance at this wavelength than
does dsDNA.] Because there are three hydrogen bonds between G and C but only
two between A and T, DNA that contains high concentrations of A and T denatures
at a lower temperature than G-and C-rich DNA (Figure 29.6). Under appropriate
conditions, complementary DNA strands can reform the double helix by the
process called renaturation (or reannealing). [Note: Separation of the two
strands over short regions occurs during both DNA and RNA synthesis.]
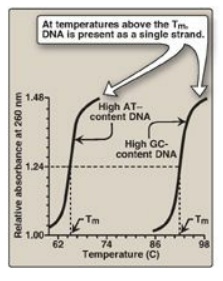
Figure 29.6 Melting temperatures (Tm) of DNA molecules with different nucleotide compositions.
3. Structural forms of the double helix: There are three major structural
forms of DNA: the B form (described by Watson and Crick in 1953), the A form,
and the Z form. The B form is a right-handed helix with 10 base pairs per 360°
turn (or twist) of the helix, and with the planes of the bases perpendicular to
the helical axis. Chromosomal DNA is thought to consist primarily of B-DNA
(Figure 29.7 shows a space-filling model of B-DNA). The A form is produced by
moderately dehydrating the B form. It is also a right-handed helix, but there
are 11 base pairs per turn, and the planes of the base pairs are tilted 20°
away from the perpendicular to the helical axis. The conformation found in DNA-RNA
hybrids or RNA-RNA double-stranded regions is probably very close to
the A form. Z-DNA is a left-handed helix that contains 12 base pairs per turn
(see Figure 29.7). [Note: The deoxyribose– phosphate backbone “zigzags,” hence,
the name “Z”-DNA.] Stretches of Z-DNA can occur naturally in regions of DNA
that have a sequence of alternating purines and pyrimidines (for example, poly
GC). Transitions between the B and Z helical forms of DNA may play a role in
regulating gene expression.
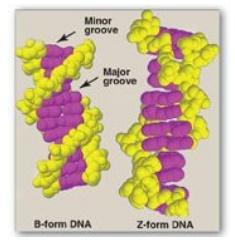
Figure 29.7 Structures of
B-DNA and Z-DNA.
C. Linear and circular DNA molecules
Each chromosome in the
nucleus of a eukaryote contains one long, linear molecule of dsDNA, which is
bound to a complex mixture of proteins (histone and nonhistone) to form
chromatin. Eukaryotes have closed, circular, dsDNA molecules in their
mitochondria, as do plant chloroplasts. A prokaryotic organism typically
contains a single, circular, dsDNA molecule. [Note: Circular DNA is
“supercoiled”, that is, the double helix crosses over on itself one or more
times. Supercoiling can result in overwinding (positive supercoiling) or
underwinding (negative supercoiling) of DNA. Supercoiling, a type of tertiary
structure, compacts DNA.] Each prokaryotic chromosome is associated with
nonhistone proteins that help compact the DNA to form a nucleoid. In addition,
most species of bacteria also contain small, circular, extrachromosomal DNA
molecules called plasmids. Plasmid DNA carries genetic information, and
undergoes replication that may or may not be synchronized to chromosomal
division. [Note: The use of plasmids as vectors in recombinant DNA technology
is described in Chapter 33.]
Plasmids may carry genes that convey antibiotic resistance to the host bacterium and may facilitate the transfer of genetic information from one bacterium to another.
Related Topics
