Chapter Summary, Questions Answers - Regulation of Gene Expression
| Home | | Biochemistry |Chapter: Biochemistry : Regulation of Gene Expression
Gene expression results in the production of a functional gene product (either RNA or protein) through the processes of replication, transcription, and translation.
CHAPTER SUMMARY
Gene expression results
in the production of a functional gene product (either RNA or protein) through
the processes of replication, transcription, and translation (Figure 32.19).
Genes can be either constitutive (always expressed, housekeeping genes) or regulated
(expressed only under certain conditions in all cells or in a subset of cells).
The ability to appropriately express (positive regulation) or repress (negative
regulation) genes is essential in all organisms. Regulation of gene expression
occurs primarily at the level of transcription in both prokaryotes and
eukaryotes and is mediated through the binding of trans-acting proteins to
cis-acting regulatory elements on the DNA. In eukaryotes, regulation also
occurs through modifications to the DNA as well as through posttranscriptional
a nd posttranslational events. In prokaryotes, such as E. coli, the coordinate
regulation of genes whose protein products are required for a particular
process is achieved through operons (groups of genes sequentially arranged on
the chromosome along with the regulatory elements that determine their
transcription). The lac operon contains the Z, Y, and A structural genes, the
protein products of which are needed for the catabolism of lactose. It is
subject to negative and positive regulation. When glucose is available, the
operon is repressed by the binding of t h e repressor protein (the product of
the lacI gene) to the operator, thus preventing transcription. When only
lactose is present, the operon is induced by an isomer of lactose (allolactose)
that binds the repressor protein, preventing it from binding to the operator.
In addition, cyclic AMP binds the catabolite-activator protein (CAP), and the
complex binds the DNA at the CAP site. This increases promoter efficiency and
results in the expression of the structural genes through the production of a
polycistronic messenger RNA (mRNA). When both glucose and lactose are present,
glucose prevents formation of cAMP and transcription of these genes is
negligible. The trp operon contains genes needed for the synthesis of
tryptophan (Trp), and, like the lac operon, it is regulated by negative
control. Unlike the lac operon, it is also regulated by attenuation, in which
mRNA synthesis that escaped repression by Trp is terminated before completion.
Transcription of ribosomal RNA and transfer RNA is selectively inhibited in
prokaryotes by the stringent response to amino acid starvation. Translation is
also a site of prokaryotic gene regulation: Excess ribosomal proteins bind the
Shine-Dalgarno sequence on their own polycistronic mRNA, preventing ribosomes
from binding. Gene regulation is more complex in eukaryotes. Operons typically
are not present, but coordinate regulation of the transcription of genes
located on different chromosomes can be achieved through the binding of
trans-acting proteins to cis-acting elements. In multicellular organisms,
hormones can cause coordinated regulation, either through the binding of the
hormone receptor–hormone complex to the DNA (as with steroid hormones) or
through the binding of a protein that is activated in response to a second
messenger (as with glucagon). In each case, binding to DNA is mediated through
structural motifs such as the zinc finger. Co- and posttranscriptional
regulation is also seen in eukaryotes and includes splice-site choice,
polyA-site choice, mRNA editing, and variations in mRNA stability as seen with
transferrin receptor synthesis and with RNA interference. Regulation at the
translational level can be caused by the phosphorylation and inhibition of
eukaryotic initiation factor, eIF-2. Gene expression in eukaryotes is also
influenced by availability of DNA to the transcriptional apparatus, the amount
of DNA, and the arrangement of the DNA. Epigenetic changes to histone proteins
and DNA also influence gene expression.
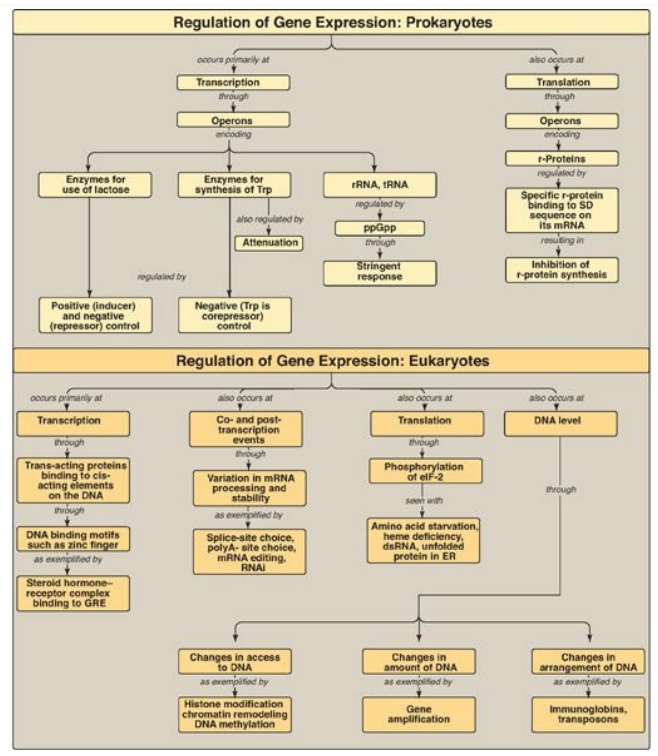
Figure 32.19 Summary of key concepts for the regulation of gene expression. GRE = glucocorticoid-response element; ppGpp = polyphosphorylated guanosine; r-protein = ribosomal protein; SD sequence = Shine-Dalgarno sequence; RNAi = RNA interference; eIF-2 = eukaryotic initiation factor 2.
Study Questions
Choose the one best answer.
32.1 Which of the following mutations is most
likely to result in reduced expression of the lac operon?
A. cyA- (no adenylyl cyclase made)
B. i– (no repressor
protein made)
C. Oc (operator cannot
bind repressor protein)
D. One resulting in
functionally impaired glucose transport
Correct answer = A. In the absence of glucose, adenylyl cyclase makes cyclic AMP, which forms a complex with the catabolite activator protein (CAP). The cAMP–CAP complex binds the CAP site on the DNA, causing RNA polymerase to bind more efficiently to the lac operon promoter, thereby increasing expression of the operon. With cyA- mutations, adenylyl cyclase is not made, and so the operon is unable to be turned on even when glucose is absent and lactose is present. The absence of a repressor protein or decreased ability of the repressor to bind the operator results in constitutive (constant) expression of the lac operon.
32.2 Which of the following is best described as
cis acting?
A. Cyclic AMP response
element-binding protein
B. Operator
C. Repressor protein
D. Thyroid hormone
nuclear receptor
Correct answer = B. The operator is part of the DNA
itself, and so is cis acting. The cAMP response element-binding protein,
repressor protein, and thyroid hormone nuclear receptor protein are molecules
that transit to the DNA, bind, and affect the expression of that DNA and so are
trans acting.
32.3 Which of the following is the basis for the
intestine-specific expression of apolipoprotein B-48?
A. DNA rearrangement
and loss
B. DNA transposition
C. RNA alternative
splicing
D. RNA editing
E. RNA interference
Correct answer = D. The production of apolipoprotein
(apo) B-48 in the intestine and apoB-100 in liver is the result of RNA editing
in the intestine, where a sense codon is changed to a nonsense codon by
posttranscriptional deamination of cytosine to uracil. DNA rearrangement and
transposition, as well as RNA interference and alternate splicing, do alter
gene expression but are not the basis of apoB-48 tissue-specific production.
32.4 Which of the following is most likely to be
true in hemochromatosis, a disease of iron accumulation?
A. The mRNA for the
transferrin receptor (TfR) is stabilized by the binding of iron regulatory
proteins to 3 iron-responsive elements.
B. The mRNA for the
transferrin receptor is not bound by iron regulatory proteins and is degraded.
C. The mRNA for
apoferritin is not bound by iron regulatory proteins at its 5 iron-responsive
element and is translated.
D. The mRNA for
apoferritin is bound by iron regulatory proteins and is not translated.
E. Both B and C are correct.
Correct answer = E. When iron levels in the body are
high, as is seen with hemochromatosis, there is increased synthesis of the
iron-storage molecule, apoferritin, and decreased synthesis of the transferrin
receptor (TfR) that mediates iron uptake by cells. These effects are the result
of trans-acting iron regulatory proteins binding cis-acting iron-responsive
elements, resulting in degradation of the mRNA for TfR, and increased
translation of the mRNA for apoferritin.
32.5 Patients with estrogen receptor–positive
(hormone responsive) breast cancer may be treated with the drug tamoxifen,
which binds the estrogen nuclear receptor without activating it. Which of the
following is the most logical outcome of tamoxifen use?
A. Increased
acetylation of estrogen-responsive genes
B. Increased growth of
estrogen receptor–positive breast cancer cells C. Increased production of
cyclic AMP D. Inhibition of the estrogen operon
E. Inhibition of transcription of
estrogen-responsive genes
Correct answer = E. Tamoxifen competes with estrogen
for binding to the estrogen nuclear receptor. Tamoxifen fails to activate the
receptor, preventing its binding to DNA sequences that upregulate expression of
estrogen-responsive genes. Tamoxifen, then, blocks the growth-promoting effects
of these genes and results in growth inhibition of estrogen-dependent breast
cancer cells. Acetylation increases transcription by relaxing the nucleosome.
Cyclic AMP is a regulatory signal mediated by cell-surface rather than nuclear
receptors. Mammalian cells do not have operons.
32.6 The ZYA region of the lac operon will be
maximally expressed if:
A. cyclic AMP levels
are low.
B. glucose and lactose
are both available.
C. the attenuation
stem–loop is able to form.
D. the CAP site is occupied.
Correct answer = D. It is only when glucose is gone,
cyclic AMP levels are increased, the cAMP–catabolite activator protein (CAP)
complex is bound to the CAP site, and lactose is available that the operon is
maximally expressed (induced). If glucose is present, the operon is off as a
result of catabolite repression. The lac operon is not regulated by
attenuation, a mechanism for decreasing transcription in some operons such as
the tryptophan operon.
32.7 X chromosome inactivation is a process by
which one of two X chromosomes in human females is condensed and inactivated to
prevent overexpression of X-linked genes. What would most likely be true about
the degree of DNA methylation and histone acetylation on the inactivated X
chromosome?
Cytosines in CG islands
would be hypermethylated and histone proteins would be deacetylated. Both
conditions are associated with decreased gene expression, and both are
important in maintaining X inactivation.
Related Topics
