Regulation of Eukaryotic Gene Expression
| Home | | Biochemistry |Chapter: Biochemistry : Regulation of Gene Expression
The higher degree of complexity of eukaryotic genomes, as well as the presence of a nuclear membrane, necessitates a wider range of regulatory processes.
REGULATION OF EUKARYOTIC GENE EXPRESSION
The higher degree of complexity of eukaryotic genomes, as well as the presence of a nuclear membrane, necessitates a wider range of regulatory processes. As with the prokaryotes, the primary site of regulation is at the level of transcription. Again, the theme of trans-acting molecules binding to cis-acting elements is seen. Operons, however, are generally not found in eukaryotes, which must use alternative strategies to solve the problem of how to coordinately regulate all the genes required for a specific response. In eukaryotes, gene expression is also regulated at multiple levels other than transcription. For example, the major modes of posttranscriptional regulation at the mRNA level are alternative mRNA splicing, control of mRNA stability, and control of translational efficiency. Additional regulation at the protein level occurs by mechanisms that modulate stability, processing, or targeting of the protein.
A. Trans-acting molecules
Specific transcription
factors are trans-acting DNA-binding proteins that function as transcriptional
activators. They have at least two binding domains: the DNA-binding domain, and
the transcription-activation domain. The DNA-binding domain contains specific
structural motifs, such as zinc fingers, that bind consensus sequences in DNA.
The transcription-activation domain recruits other proteins, such as the
general transcription factors ([GTFs]) and coactivators (for example, histone
acetyltransferases [HATs];). These facilitate formation of the transcription
initiation complex (RNA polymerase II plus the GTFs) at the promoter, and,
thus, activate transcription (Figure 32.9). Regulation is achieved by the
formation of a multiprotein complex bound to DNA, with protein–protein and
protein–DNA interactions controlling assembly of the complex. Although
activation domains recruit a variety of proteins, the specific effect of any
one of them is dependent upon the protein composition of the complex. This is
known as combinatorial control. [Note: DNA-binding proteins can also inhibit
transcription.]
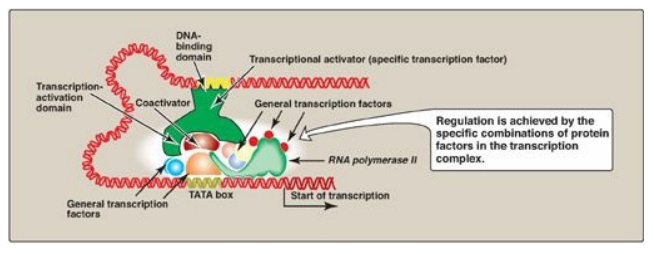
Figure 32.9 Combinatorial control of transcription.
B. Cis-acting regulatory elements
The need to coordinately regulate a group of genes to cause a particular response is of key importance in multicellular organisms including humans. An underlying theme occurs repeatedly: A protein binds to a regulatory consensus element on each of the genes in the group and coordinately affects the expression of those genes, even if they are on different chromosomes. For example, hormone-response elements (HREs) are cis-acting DNA sequences that bind trans-acting protein factors and regulate gene expression in response to hormonal signals. In general, hormones bind either to intracellular receptors (steroid hormones are an example;) or to cell-surface receptors (the peptide hormone glucagon is an example;).
1. Regulatory signals mediated by intracellular receptors: Members of the nuclear receptor superfamily, which includes the steroid hormone (glucocorticoids, mineralocorticoids, androgens, and estrogens), vitamin D, retinoic acid, and thyroid hormone receptors, all directly influence gene expression by functioning as specific transcription factors. These receptors, therefore, contain a DNA-binding domain and an activation domain. They also contain a ligand-binding domain. For example, steroid hormones such as cortisol (a glucocorticoid) bind to soluble, intracellular receptors at the ligand-binding domain (Figure 32.10). Binding causes a conformational change in the receptor that activates it. The receptor–ligand complex enters the nucleus, dimerizes, and binds via a zinc finger motif to nuclear DNA at a cis-acting regulatory element, the glucocorticoid-response element (GRE), an example of an HRE. Binding allows recruitement of coactivators to the activation domain and results in increased expression of cortisol-responsive genes, each of which is under the control of its own GRE. Binding of the receptor–hormone complex to the GRE allows coordinate expression of a group of target genes, even when these genes are located on different chromosomes. The GRE can be located upstream or downstream of the genes it regulates and is able to function at great distances from those genes. The GRE, then, can function as a true enhancer. [Note: If associated with corepressors, hormone–receptor complexes inhibit transcription.]
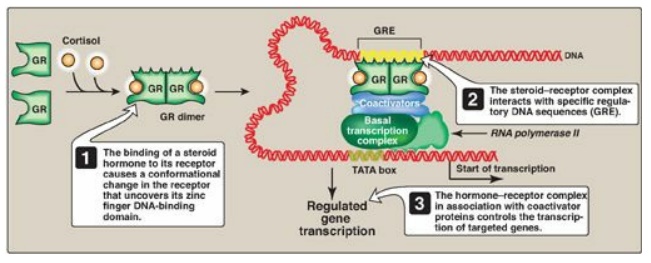
Figure 32.10 Transcriptional regulation by intracellular steroid hormone receptors. GRE = glucocorticoid-response element (an example of a hormone-response element); GR = glucocorticoid receptor.
2. Regulatory signals mediated by cell-surface
receptors:
Cell-surface receptors include those for insulin, epinephrine, and glucagon.
Glucagon, for example, is a peptide hormone that binds its G protein–coupled
plasma membrane receptor on glucagon-responsive cells. This extracellular
signal is then transduced to intracellular cAMP (Figure 32.11; also see Figure
8.7), which can affect protein expression (and activity) through protein kinase
A-mediated phosphorylation. In response to a rise in cAMP, a
trans-acting factor (cAMP response element–binding [CREB] protein) is
phosphorylated and activated. Active CREB protein binds via a leucine zipper
motif to a cis-acting regulatory element, the cAMP response element (CRE),
resulting in transcription of target genes with CREs in their promoters. [Note:
The genes for phosphoenolpyruvate carboxykinase and glucose 6-phosphatase, key
enzymes of gluconeogenesis, are examples of genes upregulated by the
cAMP/CRE/CREB system.]
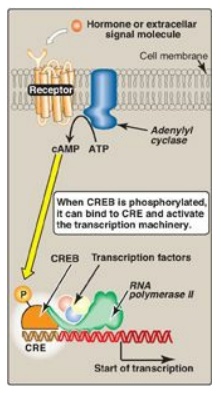
Figure 32.11 Transcriptional regulation by receptors located in the cell membrane. [Note: Cyclic AMP activates protein kinase A that phosphorylates cAMP response element-binding (CREB) protein.] CRE = cAMP response element.
C. Regulation by processing of messenger RNA
Eukaryotic mRNA
undergoes several modifications before it is exported from the nucleus to the
cytoplasm for use in protein synthesis. Capping at the 5I - end,
polyadenylation at the 3I -end, and splicing are essential processing events
for the production of a functional eukaryotic messenger from most pre-mRNA, and
variations in these events can affect gene expression. In addition, messenger
stability also affects gene expression.
1. Splice-site choice: Tissue-specific protein isoforms
can be made from the same pre-mRNA through differential cotranscriptional processing,
particularly the use of alternative splice sites (Figure 32.12). For example,
tropomyosin (TM) is an actin filament–binding protein that regulates the
functions of actin in both muscle and nonmuscle cells. Its pre-mRNA undergoes
tissue-specific differential splicing to yield a number of TM isoforms.
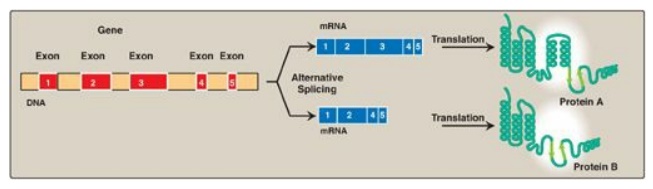
Figure 32.12 Tissue-specific alternative splicing produces multiple related proteins, or isoforms, from a single gene.
Over 60% percent of the approximately 25,000 genes
in the human genome undergo differential splicing. The use of alternative
polyadenylation and transcription start sites is also seen in many genes. This
explains, at least in part, how 25,000 genes can give rise to hundreds of
thousands of proteins.
2. Messenger RNA editing: Even after mRNA has been fully
processed, it may undergo additional posttranscriptional modification in which
a base in the mRNA is altered. This is known as RNA editing. An important
example in humans occurs with the transcript for apolipoprotein (apo) B, an
essential component of chylomicrons and very low density lipoproteins. Apo B
mRNA is made in the liver and the small intestine. However, in the intestine
only, the C residue in the CAA codon for glutamine is deaminated to U, changing
the sense codon to a nonsense or stop codon (UAA), as shown in Figure 32.13.
This results in a shorter protein (apo B-48, representing 48% of the message)
being made in the intestine (and incorporated into chylomicrons) than is made
in the liver (apo B-100, full-length, incorporated into VLDL).
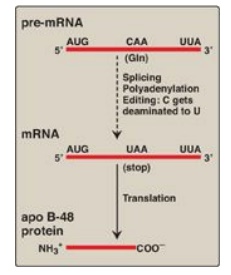
Figure 32.13 RNA editing of
apolipoprotein (apo) B in the intestine and generation of the apo B-48 protein
needed for chylomicron synthesis. Gln = glutamine; mRNA = messenger RNA.
3. Messenger RNA stability: How long an mRNA remains in the
cytosol before it is degraded influences how much protein product can be
produced from it. Regulation of iron metabolism and the gene-silencing process
of RNA interference illustrate the importance of mRNA stability in the
regulation of gene expression.
a. Iron metabolism: Transferrin is a plasma protein
that transports iron. Transferrin binds to cell-surface receptors (transferrin
receptors [TfRs]) that get internalized and provide cells, such as
erythroblasts, with iron. The mRNA for the TfR has several cis-acting
iron-responsive elements (IREs) at its 3-end. IREs have a short stem–loop
structure that can be bound by trans-acting iron regulatory proteins ([IRPs]
Figure 32.14). When the iron concentration in the cell is low, the IRPs bind to
the 3-IREs and stabilize the mRNA for TfR, allowing TfR synthesis. When
intracellular iron levels are high, the IRPs are degraded. The lack of IRPs
bound to the mRNA hastens its destruction, resulting in decreased TfR
synthesis. [Note: The mRNA for apoferritin, an intracellular protein of iron
storage, has a single IRE at its 5I -end. When iron levels in the cell are low,
IRPs bind the 5I -IRE and prevent the use of the mRNA, and less apoferritin is
made. When iron accumulates in the cell, the IRP is degraded, allowing
synthesis of apoferritin molecules to store the excess iron. ALAS2, the
regulated enzyme of heme synthesis in erythroblasts, also contains a 5-IRE.]
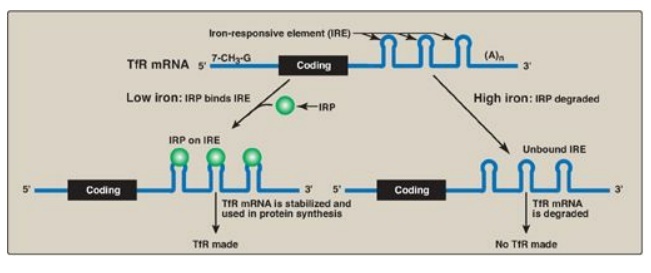
Figure 32.14 Regulation of
transferrin receptor (TfR) synthesis. IRP = iron regulatory protein. [Note: The
IREs are located in the 3I UTR (untranslated region) of the TfR mRNA.]
b. RNA interference: RNA interference (RNAi) is a
mechanism of gene silencing through decreased expression of mRNA, either by
repression of translation or by increased degradation. It is thought to play a
key role in such fundamental processes as cell proliferation, differentiation,
and apoptosis. RNAi is mediated by short (~22 bp), noncoding RNAs called
microRNAs (miRNAs), which arise from far longer, genomically encoded nuclear
transcripts, primary miRNA (pri-miRNA) that are partially processed in the
nucleus to pre-miRNA by an endonuclease (Drosha) then transported to the
cytoplasm. There, an endonuclease (Dicer) completes the processing and
generates short, double-stranded miRNA. A single strand (the guide, or
antisense strand) of the miRNA associates with a cytosolic protein complex
known as the RNA-induced silencing complex (RISC). The guide strand hybridizes
with a complementary sequence on a full length target mRNA, bringing RISC to
the mRNA. This can result in repression of translation of the mRNA or its degradation
by an endonuclease (Argonaute/Ago/Slicer) of the RISC. The extent of
complementarity appears to be the determining factor (Figure 32.15). RNAi can
also be triggered by double-stranded short interfering RNAs (siRNAs) introduced
into a cell from exogenous sources. [Note: In vertebrates, the function of
siRNAs that may arise from endogenous sources is unclear.]
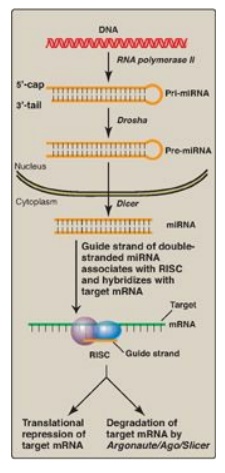
Figure 32.15 Biogenesis and
actions of miRNA. [Note: The extent of complementarity between the target
messenger RNA (mRNA) and the microRNA (miRNA) determines the final outcome.]
RISC = RNA-induced silencing complex.
RNA-interference–based therapeutics: Modulation of gene expression by
providing siRNA to trigger RNAi has enormous therapeutic potential. The first
clinical trial of RNAi-based therapy involved patients with the neovascular
form of age-related macular degeneration (AMD), a leading form of adult
blindness. Neovascular AMD is triggered by overproduction of vascular
endothelial growth factor (VEGF), leading to the sprouting of excess blood
vessels behind the retina. The vessels leak, clouding and often entirely
destroying vision (therefore, neovascular AMD is also referred to as “wet”
macular degeneration). An siRNA designed to target the mRNA of VEGF and promote
its degradation went to clinical trials. Although considerable effort and
resources have been expended to develop RNAi-based therapeutics, especially for
the treatment of cancer, no products have gone from trials to the market.
Development has been hindered by the problems of targeted delivery and
stability. The use of nano-sized vectors such as liposomes may eliminate these
issues. The research applications of RNAi, however, have grown rapidly.
4. Translation of messenger RNA: Regulation of gene expression can
also occur at the level of translation. One mechanism by which translation is
regulated is through phosphorylation of the eukaryotic translation initiation
factor, eIF-2 ( Figure 32.16). Phosphorylation of eIF-2 inhibits its function
and so inhibits translation at the initiation step. [Note: Phosphorylation of
eIF-2 prevents its reactivation by inhibiting GDP–GTP exchange.]
Phosphorylation is catalyzed by kinases that are activated in response to
environmental conditions, such as amino acid starvation, heme deficiency in
erythroblasts, the presence of double-stranded RNA (signaling viral infection),
and the accumulation of misfolded proteins in the rough endoplasmic reticulum.
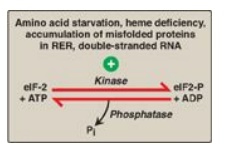
Figure 32.16 Regulation of
translation initiation in eukaryotes by phosphorylation of eukaryotic
translation initiation factor, eIF-2. RER = rough endoplasmic reticulum; ADP =
adenosine diphosphate; Pi = inorganic phosphate.
D. Regulation through modifications to DNA
Gene expression in
eukaryotes is also influenced by the availability of DNA to the transcriptional
apparatus, the amount of DNA, and the arrangement of DNA. [Note: Localized
transitions between the B and Z forms of DNA can also affect gene expression.]
1. Access to DNA: In eukaryotes, DNA is found complexed
with histone and nonhistone proteins to form chromatin. Transcriptionally
active, decondensed chromatin (euchromatin) differs from the more condensed,
inactive form (heterochromatin) in a number of ways. Active chromatin contains
histone proteins that have been covalently modified at their amino terminal
ends by acetylation or phosphorylation. Such modifications decrease the
positive charge of these basic proteins, thereby decreasing the strength of
their association with negatively charged DNA. This relaxes the nucleosome,
allowing transcription factors access to specific regions on the DNA.
Nucleosomes can also be repositioned, an ATP-requiring process called chromatin
remodeling. Another difference between transcriptionally active and inactive
chromatin is the extent of methylation of cytosine bases in CG-rich regions
(CpG islands) in the promoter region of many genes. Methylation is by
methyltransferases that use S-adenosylmethionine as the methyl donor (Figure
32.17). Transcriptionally active genes are less methylated (hypomethylated)
than their inactive counterparts, suggesting that DNA hypermethylation silences
gene expression. [Note: Modification of histones and methylation of DNA are
epigenetic. They are heritable changes in DNA that alter gene expression
without altering the base sequence.]
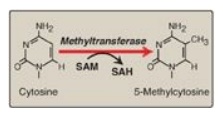
Figure 32.17 The methylation
of cytosine in eukaryotic DNA. SAM = S-adenosylmethionine; SAH =
S-adenosylhomocysteine.
2. Amount of DNA: A change up or down in the number of copies of a gene can affect the amount of gene product produced. An increase in copy number (gene amplification) has contributed to increased genomic complexity and is still a normal developmental process in certain nonmammalian species. In mammals, however, gene amplification is seen with some diseases and in response to particular chemotherapeutic drugs such as methotrexate, an inhibitor of the enzyme dihydrofolate reductase (DHFR), required for the synthesis of thymidine triphosphate (TTP) in the pyrimidine biosynthetic pathway. TTP is essential for DNA synthesis. Gene amplification results in an increase in the number of DHFR genes and resistance to the drug, allowing TTP to be made.
3. Arrangement of DNA: The process by which immunoglobulins
(antibodies) are produced by B lymphocytes involves permanent rearrangements of
the DNA in these cells. The immunoglobulins (for example, IgG) consist of two
light and two heavy chains, with each chain containing regions of variable and
constant amino acid sequence. The variable region is the result of somatic
recombination of segments within both the light- and the heavy-chain genes.
During B-lymphocyte development, single variable (V), diversity (D), and
joining (J) gene segments are brought together through gene rearrangement to
form a unique variable region (Figure 32.18). This process allows the
generation of 109–1011 different immunoglobulins from a single gene, providing
the diversity needed for the recognition of an enormous number of antigens.
[Note: The shift from the membrane-bound form to the secreted form of
immunoglobulins involves poly-A site choice.]
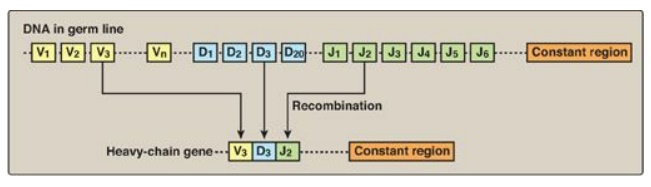
Figure 32.18 DNA rearrangements in the generation of immunoglobulins. V= variable; D = diversity; J = joining.
4. Mobile DNA elements: Transposons (Tns) are mobile segments of DNA that move in an essentially random manner from one site to another on the same or a different chromosome. Movement is mediated by transposase, an enzyme encoded by the Tn itself. Movement can be direct, in which transposase cuts out and then inserts the Tn at a new site, or replicative, in which the Tn is copied and the copy inserted elsewhere while the original remains in place. In eukaryotes, including humans, replicative transposition frequently involves an RNA intermediate, in which case the Tn is called a retrotransposon. Transposition has contributed to structural variation in the genome but also has the potential to alter gene expression and even to cause disease. Although the vast majority of retrotransposons in the human genome have lost the ability to move, some are still active. Their transposition is thought to be the basis for some rare cases of hemophilia A and Duchenne muscular dystrophy. [Note: The growing problem of antibiotic-resistant bacteria is a consequence, at least in part, of the exchange of plasmids among bacterial cells. If the plasmids contain Tns carrying antibiotic resistance genes, the recipient bacteria gain resistance to one or more antimicrobial drugs.]
Related Topics
