DNA Cloning
| Home | | Biochemistry |Chapter: Biochemistry : Biotechnology and Human Disease
Introduction of a foreign DNA molecule into a replicating cell permits the cloning or amplification (that is, the production of many identical copies) of that DNA.
DNA CLONING
Introduction of a foreign DNA molecule into a replicating cell permits the cloning or amplification (that is, the production of many identical copies) of that DNA. [Note: Human DNA for cloning can be obtained from blood, saliva, and solid tissue.] In some cases, a single DNA fragment can be isolated and purified prior to cloning. More commonly, to clone a nucleotide sequence of interest, the total cellular DNA is first cleaved with a specific restriction enzyme, creating hundreds of thousands of fragments. Each of the resulting DNA fragments is joined to a DNA vector molecule (referred to as a cloning vector) to form a hybrid, or recombinant, DNA molecule. Each recombinant molecule carries its inserted DNA fragment into a single host cell (for example, a bacterium), where it is replicated. [Note: The process of introducing foreign DNA into a cell is called transformation for bacteria and yeast and transfection for higher eukaryotes.] As the host cell multiplies, it forms a clone in which every bacterium contains copies of the same inserted DNA fragment, hence the name “cloning.” The cloned DNA can be released from its vector by cleavage (using the appropriate restriction endonuclease) and isolated. By this mechanism, many identical copies of the DNA of interest can be produced. [Note: An alternative to amplification by biologic cloning, the polymerase chain reaction (PCR)]
A. Vectors
A vector is a molecule
of DNA to which the fragment of DNA to be cloned is joined. Essential
properties of a vector include: 1) it must be capable of autonomous replication
within a host cell, 2) it must contain at least one specific nucleotide sequence
recognized by a restriction endonuclease, and 3) it must carry at least one
gene that confers the ability to select for the vector such as an antibiotic
resistance gene. Commonly used vectors include plasmids and viruses.
1. Prokaryotic plasmids: Prokaryotic organisms typically
contain single, large, circular chromosomes. In addition, most species of
bacteria also normally contain small, circular, extrachromosomal DNA molecules
called plasmids ( Figure 33.5). Plasmid DNA undergoes replication that may or
may not be synchronized to chromosomal division. Plasmids may carry genes that
convey antibiotic resistance to the host bacterium and may facilitate the
transfer of genetic information from one bacterium to another. They can be
readily isolated from bacterial cells, their circular DNA cleaved at specific sites
by restriction endonucleases, and up to 10 kb (kilobases) of foreign DNA (cut
with the same restriction enzyme) inserted. The recombinant plasmid can be
introduced into a bacterium, producing large numbers of copies of the plasmid.
The bacteria are grown in the presence of antibiotics, thus selecting for cells
containing the hybrid plasmids, which provide antibiotic resistance (Figure
33.6). Artificial plasmids are routinely constructed. An example is pRB322
(Figure 33.5), which contains an origin of replication, two antibiotic
resistance genes, and over 40 unique restriction sites. Use of plasmids is
limited by the size of the DNA that can be inserted.
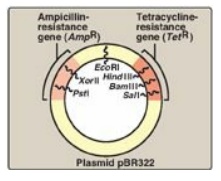
Figure 33.5 A restriction map
of plasmid pBR322 indicating the positions of its antibiotic resistance genes
and 6 of the over 40 unique sites recognized by specific restriction
endonucleases.
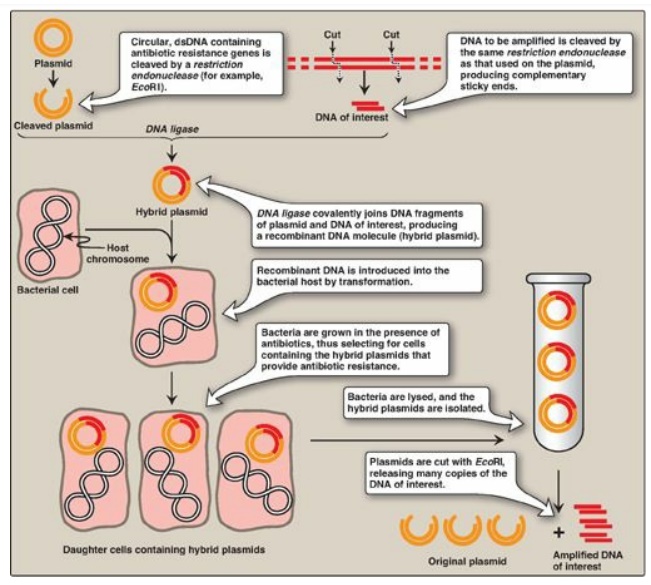
Figure 33.6 Summary of gene
cloning. dsDNA = double-stranded DNA.
2. Other vectors: The development of improved vectors that can more
efficiently accommodate larger DNA segments, or express the passenger genes in
different cell types, has aided molecular genetics research. In addition to the
prokaryotic plasmids described above, naturally occurring viruses that infect
bacteria (bacteriophage l, for example) or mammalian cells (retroviruses, for
example), as well as artificial constructs such as cosmids and bacterial or
yeast artificial chromosomes (BACs or YACs, respectively), are currently used
as cloning vectors. [Note: BACs and YACs can accept DNA inserts of 100–250 kb
and 250–1000 kb, respectively.]
B. DNA libraries
A DNA library is a
collection of cloned restriction fragments of the DNA of an organism. Two kinds
of libraries are commonly used: genomic libraries and complementary DNA (cDNA)
libraries. Genomic libraries ideally contain a copy of every DNA nucleotide
sequence in the genome. In contrast, cDNA libraries contain those DNA sequences
that only appear as processed messenger RNA (mRNA) molecules, and these differ
from one cell type to another. [Note: cDNA lacks introns and the control
regions of the genes, whereas these are present in genomic DNA.]
1. Genomic DNA libraries: A genomic library is created by digestion of the total DNA of an organism with a restriction endonuclease and subsequent ligation to an appropriate vector. The recombinant DNA molecules replicate within host bacteria. Thus, the amplified DNA fragments collectively represent the entire genome of the organism and are called a genomic library. Regardless of the restriction enzyme used, the chances are rather good that the gene of interest contains more than one restriction site recognized by that enzyme. If this is the case, and if the digestion is allowed to go to completion, the gene of interest is fragmented (that is, it is not contained in any one clone in the library). To avoid this usually undesirable result, a partial digestion is performed in which either the amount or the time of action of the enzyme is limited. This results in cleavage occurring at only a fraction of the restriction sites on any one DNA molecule, thus producing fragments of about 20 kb. Enzymes that cut very frequently (that is, those that recognize four-bp sequences) are generally used for this purpose so that the result is an almost random collection of fragments. This ensures a high degree of probability that the gene of interest is contained, intact, in some fragment.
2. Complementary DNA libraries: If a protein-coding gene of
interest is expressed at a high level in a particular tissue, the mRNA
transcribed from that gene is likely also present at high concentrations in the
cells of that tissue. For example, reticulocyte mRNA is composed largely of
molecules encoding the α-globin and b-globin chains of hemoglobin. This mRNA
can be used as a template to make a cDNA molecule using the enzyme reverse
transcriptase (Figure 33.7). The resulting cDNA is, therefore, a
double-stranded copy of mRNA. [Note: The template mRNA is isolated from
transfer RNA and ribosomal RNA by the presence of its poly-A tail.] cDNA can be
amplified by cloning or by PCR. It can be used as a probe to locate the gene
that coded for the original mRNA (or fragments of the gene) in mixtures
containing many unrelated DNA fragments. If the mRNA used as a template is a
mixture of many different size species, the resulting cDNA is heterogeneous.
These mixtures can be cloned to form a cDNA library. Because cDNA has no
intervening sequences, it can be cloned into an expression vector for the
synthesis of eukaryotic proteins by bacteria (Figure 33.8). These special
plasmids contain a bacterial promoter for transcription of the cDNA and a
Shine-Dalgarno (SD) sequence ( that allows the bacterial ribosome to initiate
translation of the resulting mRNA molecule. The cDNA is inserted downstream of
the promoter and within a gene for a protein that is expressed in the bacterium
(for example, lacZ;), such that the mRNA produced contains an SD sequence, a
few codons for the bacterial protein, and all the codons for the eukaryotic
protein. This allows for more efficient expression and results in the
production of a fusion protein. [Note: Therapeutic human insulin is made in
bacteria through this technology. However, the extensive co- and
posttranslational modifications required for most other human proteins (for
example, blood clotting factors) necessitates the use of eukaryotic, even
mammalian, hosts.]
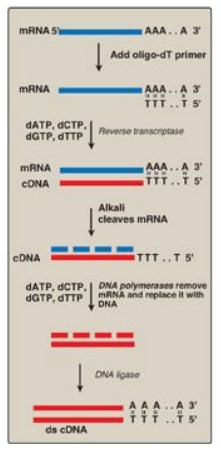
Figure 33.7 Synthesis of cDNA from messenger RNA (mRNA) using reverse transcriptase. Additional steps (not shown) are required to clone the cDNA. [Note: Recall that DNA is resistant to alkaline hydrolysis.]
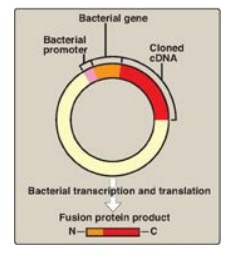
Figure 33.8 An expression
vector. The complementary DNA (cDNA) is inserted within a bacterial gene,
downstream of the promoter sequence, and the sequences for the messenger RNA
Shine-Dalgarno sequence, start codon, and codons for the first few amino acids
of the bacterial protein. The product is a fusion protein |||| that contains just some
amino acids of the bacterial protein and all the amino acids of the
cDNA-encoded protein ||| .
C. Sequencing of cloned DNA fragments
The base sequence of
DNA fragments that have been cloned can be determined. The original procedure
for this purpose was the Sanger dideoxy method illustrated in Figure 33.9. In
this method, the single-stranded DNA (ssDNA) to be sequenced is used as the
template for DNA synthesis by DNA polymerase. A radiolabeled primer
complementary to the 3I -end of the target DNA is added, along with the four
deoxyribonucleoside triphosphates (dNTPs). The sample is divided into four
reaction tubes, and a small amount of one of the four dideoxyribonucleoside
triphosphates (ddNTPs) is added to each tube. Because it contains no 3I
-hydroxyl group, incorporation of a ddNMP terminates elongation at that point. The
products of this reaction, then, consist of a mixture of DNA strands of
different lengths, each terminating at a specific base. Separation of the
various DNA products by size using polyacrylamide gel electrophoresis, followed
by autoradiography, yields a pattern of bands from which the DNA base sequence
can be read. [Note: The shorter the fragment, the farther it travels on the
gel, with the shortest fragment representing that which was made first (that
is, the 5I -end).] In place of a labeled primer, a mixture of the four ddNTPs
linked to different fluorescent dyes and in a single reaction tube is now
commonly used. [Note: The Human Genome Project used highly automated variations
of this technique to determine the base sequence of the human genome.] Advances
in sequencing technology, so-called next generation, or deep sequencing, now
allow the sequencing of longer segments in a shorter time with increased
fidelity and decreased cost through the simultaneous (parallel) sequencing of
many DNA pieces.
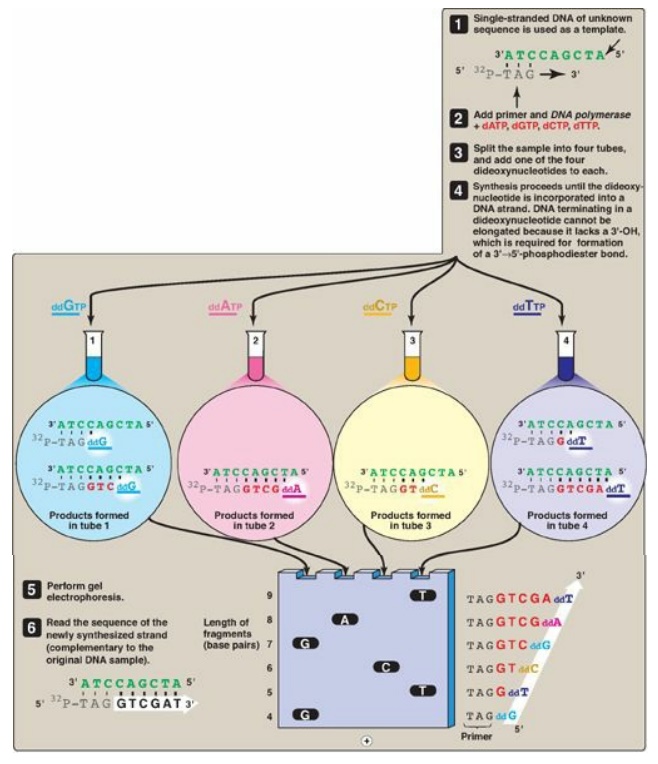
Figure 33.9 DNA sequencing by
the Sanger dideoxy method. [Note: The original method utilized a radiolabeled
primer. Fluorescent dye-labeled ddNTPs are now commonly used.] A = adenine; C =
cytosine; G = guanine; T = thymine; d = deoxy; dd = dideoxy.
Related Topics
